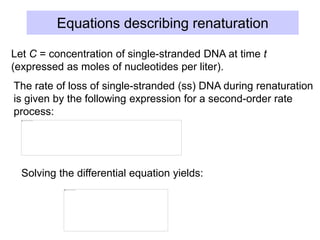
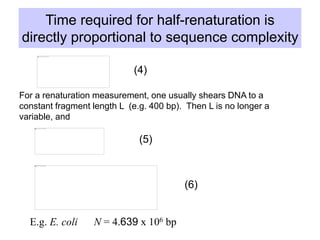
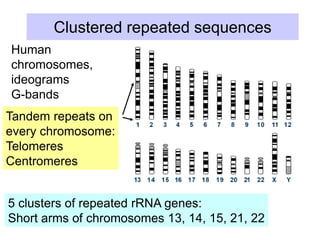
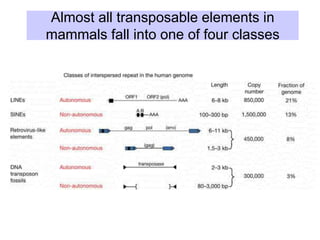
This document discusses various components that make up genomes. It describes how genomes contain both coding and non-coding DNA sequences. The non-coding portions include repetitive elements like short interspersed elements (SINEs), long interspersed elements (LINEs), endogenous retroviruses, and DNA transposons. Genome complexity is measured using DNA renaturation kinetics, where more complex genomes with greater unique sequences renature more slowly. Comparative genomics and identifying repetitive elements helps characterize genome structure and functional elements.