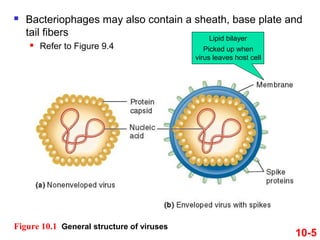
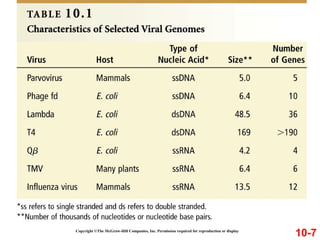
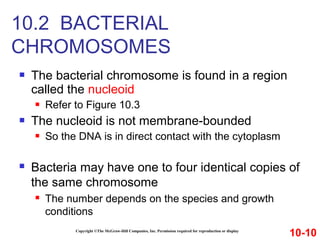
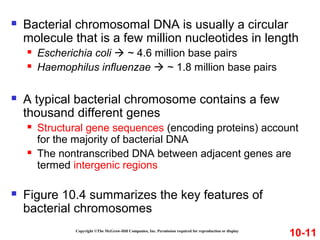
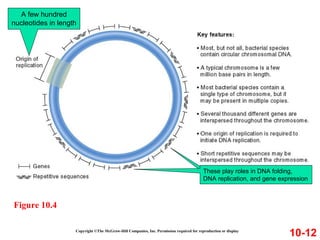
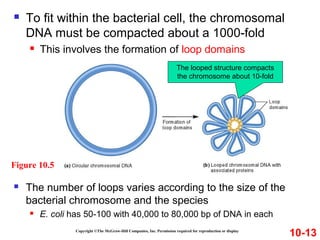
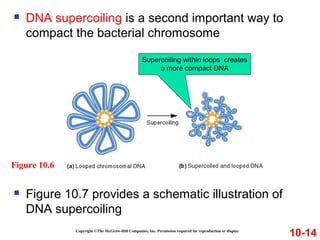
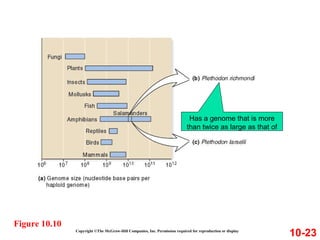
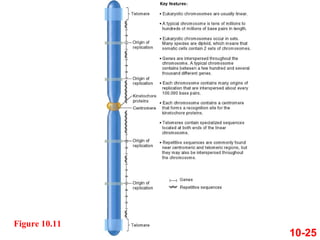
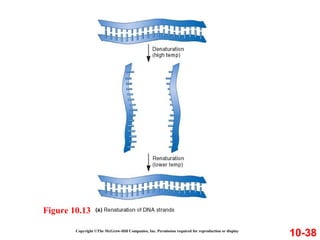
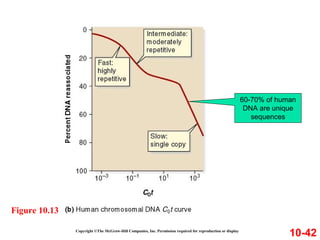
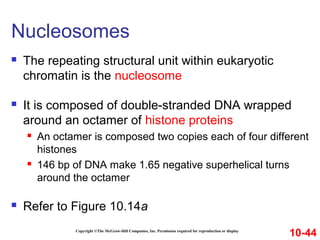
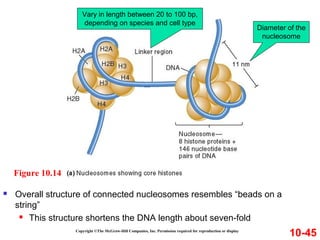
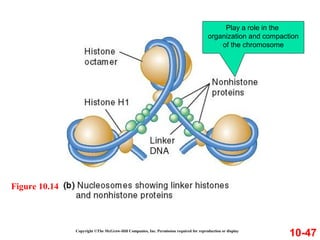
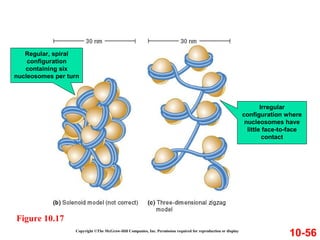
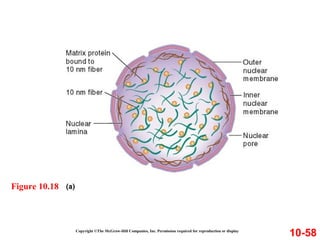
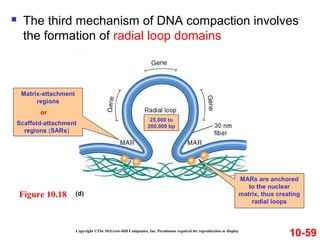
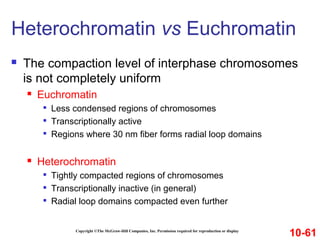
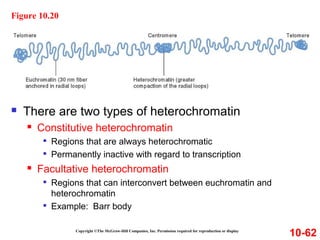
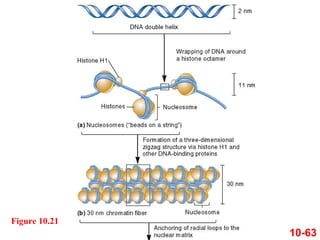
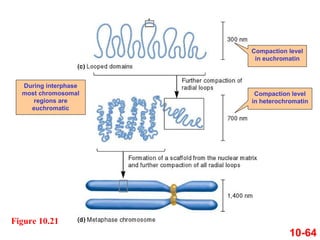
Chromosomes contain DNA and proteins. In eukaryotes, chromosomes are located in the nucleus and must be highly compacted to fit by binding proteins to form chromatin. Chromatin is compacted in a hierarchical manner through interactions with histone proteins to form nucleosomes, which further compact to form the 30nm fiber and loop domains that attach to the nuclear matrix, compacting the DNA over 1000-fold to fit in the cell.