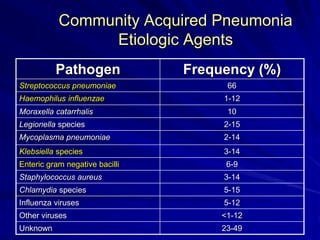
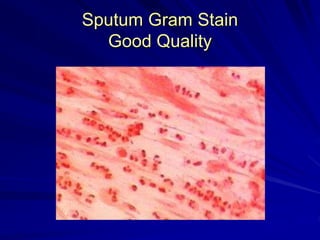
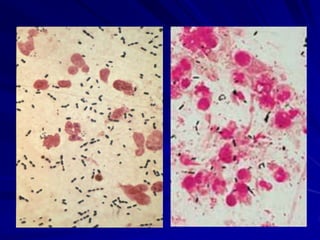
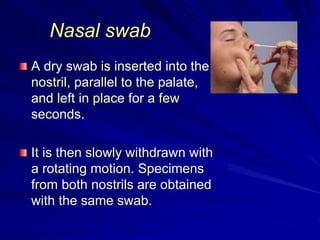
The document outlines the laboratory diagnosis of respiratory infections, detailing the common pathogens and methodologies for specimen collection and processing for both lower and upper respiratory tract infections. It emphasizes the importance of accurate specimen handling and the interpretation of results, highlighting specific tests like sputum cultures and blood cultures. Additionally, it discusses various causative agents, including bacteria, fungi, and viruses, along with their identification techniques.