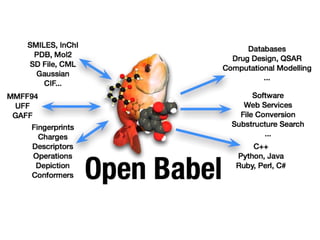
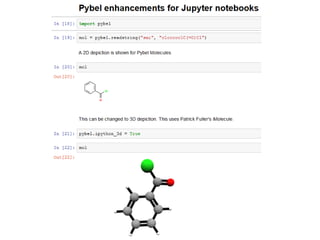
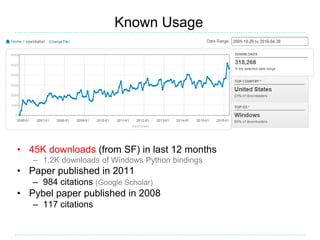
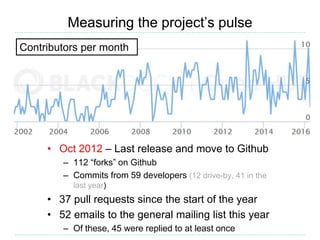
Open Babel is a chemical toolbox that includes a programming library in C++ for reading and writing many chemical file formats as well as generating 2D and 3D coordinates. It has a long history beginning in 1992 and is widely used, with over 45,000 downloads in the last year. It supports many features for cheminformatics and materials science. The code is open source under the GPL license and has contributors from both academia and industry.







![Formats: most recent additions
• Siesta [read]
– ab initio molecular dynamics
• STL [write]
– (STereoLithography) 3D
printing
• Point cloud format [write]
– Write VdW surface as points
• AOForce [read]
– Turbomole vibrational freqs
• MDFF [read/write]
– MD fitting to density maps
• EXYZ [read/write]
– Extended XYZ
git log --pretty=oneline --name-status | grep "^A" | grep src/formats | grep -v inchi | grep -v
libxml | less](https://image.slidesharecdn.com/openbabelmioss2016-160519075344/85/Open-Babel-project-overview-16-320.jpg)
![Formats: most recent additions
• Siesta [read]
– ab initio molecular dynamics
• STL [write]
– (STereoLithography) 3D
printing
• Point cloud format [write]
– Write VdW surface as points
• AOForce [read]
– Turbomole vibrational freqs
• MDFF [read/write]
– MD fitting to density maps
• EXYZ [read/write]
– Extended XYZ
git log --pretty=oneline --name-status | grep "^A" | grep src/formats | grep -v inchi | grep -v
libxml | less
• Orca [read/write]
– QM package
• JSON formats [read/write]
– ChemDoodle JSON
– PubChem JSON
• Confab report [write]
– Conformation generation
• Dalton [read]
– QM package
• LPMD [read/write]
– MD with interatomic potentials
• Smiley [read]
– Validating SMILES parser](https://image.slidesharecdn.com/openbabelmioss2016-160519075344/85/Open-Babel-project-overview-17-320.jpg)