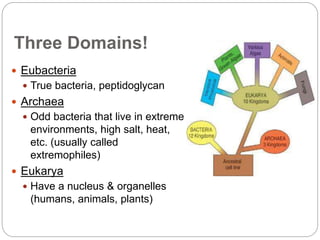
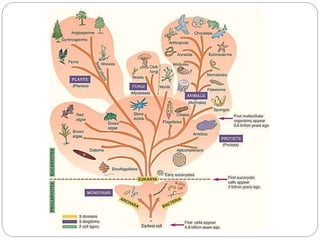
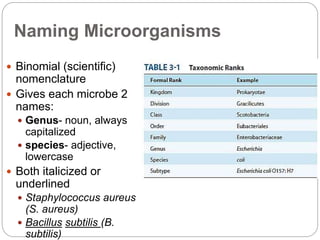
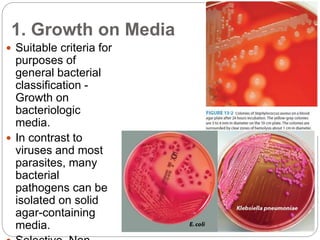
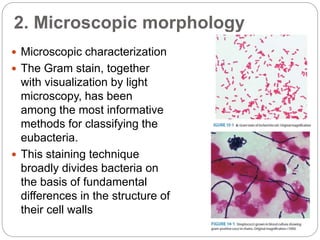
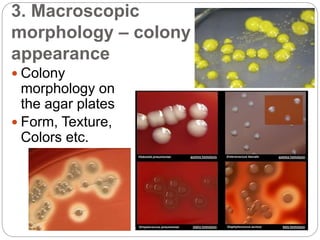
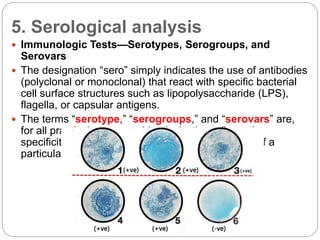
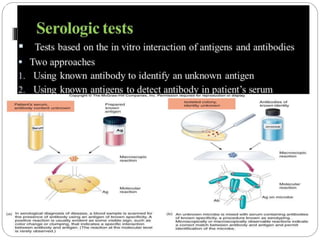
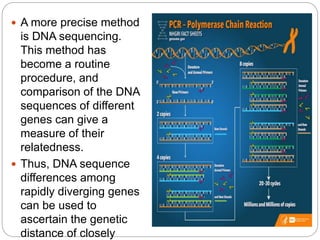
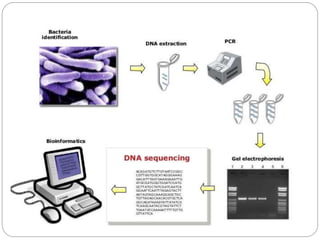
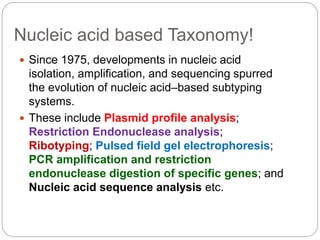
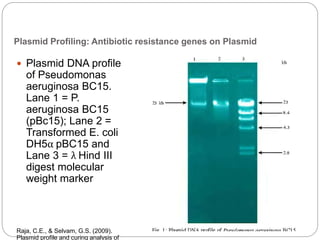
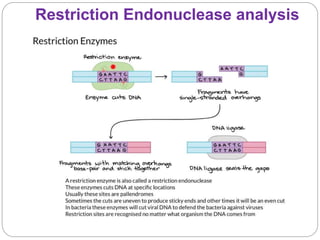
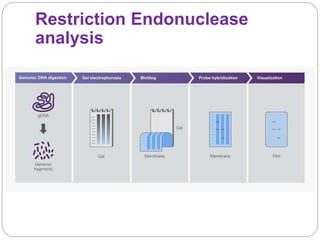
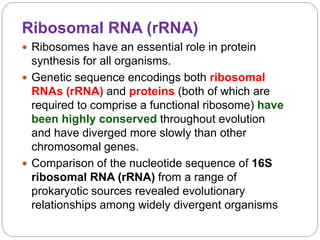
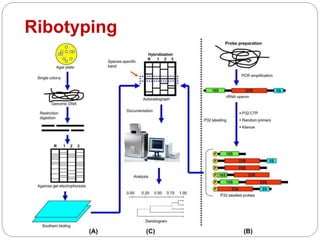
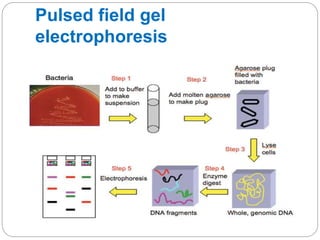
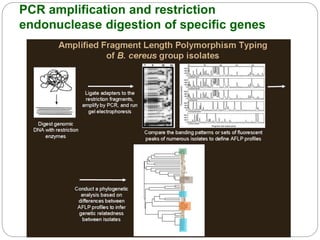
This document discusses the taxonomy and classification of microorganisms. It begins by outlining the objectives of discussing taxonomy, naming, and the classification basis for prokaryotes. It then explains Carl von Linné's development of the formal taxonomy system and how organisms are classified from domain to species. The key methods for classifying microorganisms are described as growth on media, microscopic and macroscopic morphology, biochemical tests, serological analysis, and genetic/molecular analysis like DNA sequencing. The three domains of life and binomial nomenclature for naming species are also outlined.