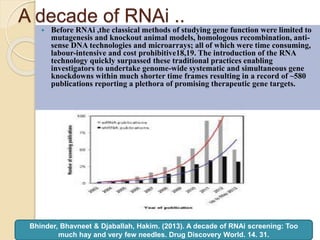
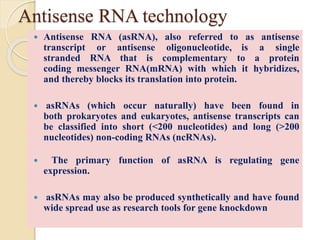
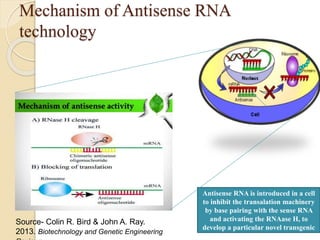
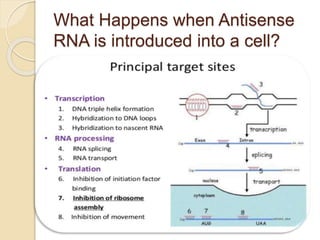
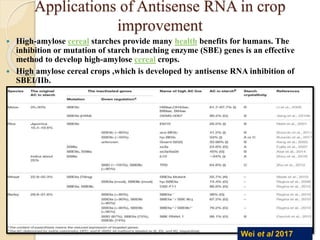
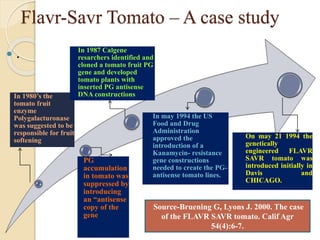
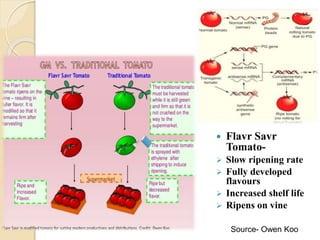
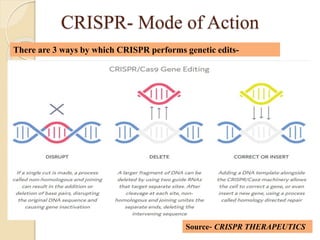
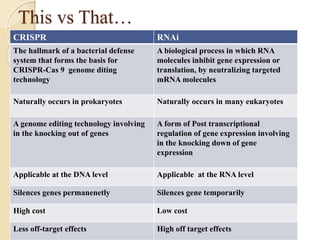
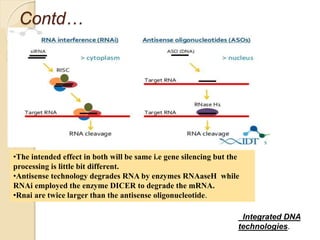
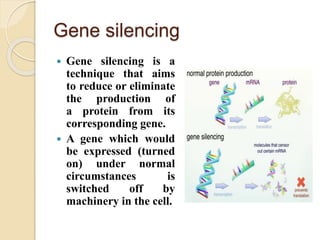
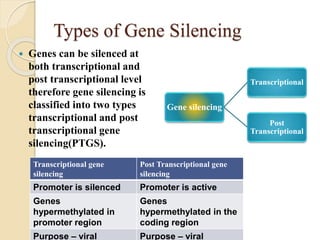
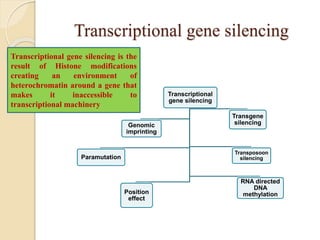
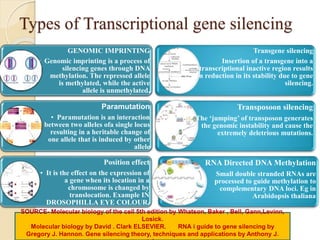
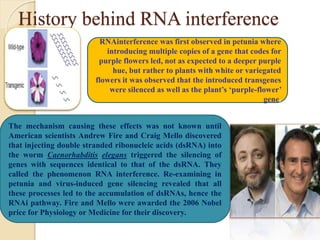
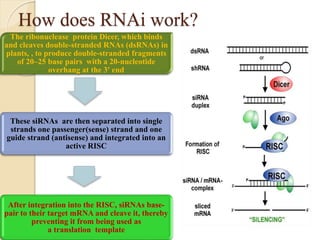
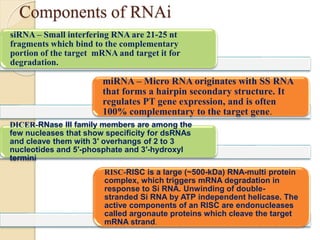
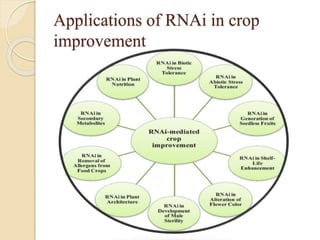
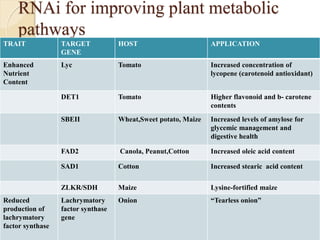
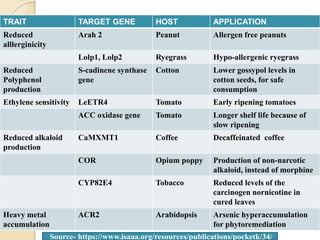
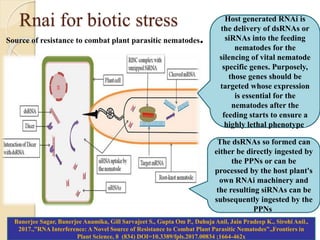
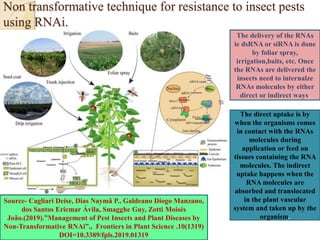
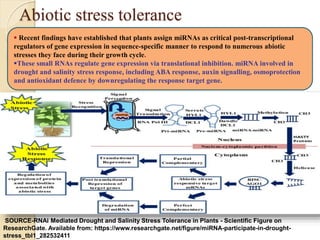
The document discusses gene silencing, a crucial technique in biotechnology for crop improvement, detailing its types, mechanisms, and applications. It emphasizes RNA interference (RNAi) as a prominent method that allows for targeted gene suppression, thus enabling advancements in crop traits and resistance to diseases and pests. The document also mentions historical discoveries related to RNAi and applications in increasing agricultural yields and enhancing crop resilience to abiotic and biotic stresses.









![Generation of transgenic cotton
(Gossypium hirsutum cv.
Narasimha) plants expressing
intergenic region [Cotton leaf curl
Rajasthan virus genome
(GQ220850)]-derived ihpRNAi
construct. Germination of seed of
cotton (G. hirsutum cv. Narasimha)
(a, b); explants on co-cultivation medium (c);
explants on selection medium (d–f);
hardening and acclimatization of plantlets in
Hoagland medium, agropeat and
glasshouse, respectively (g–i)
Whitefly (Bemisia
tabaci)-mediated
inoculation of
transgenic cotton
(Gossypium hirsutum
cv. Narasimha) plants
with Cotton leaf curl
virus. Transformed plants did
not show disease symptoms
(a); symptom development on
non-transformed cotton
showing vein thickening and
leaf curling (b)
Source - Khatoon, Sameena et al (2016).](https://image.slidesharecdn.com/jhilickcreditseminar-210628075946/85/Gene-silencing-techniques-for-crop-improvement-26-320.jpg)