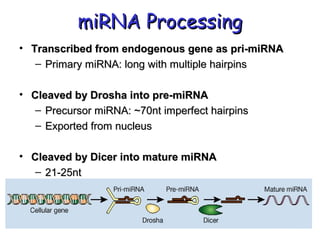
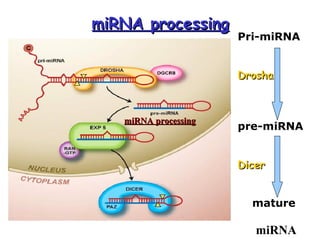
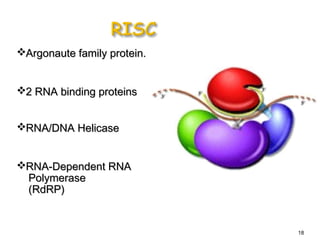
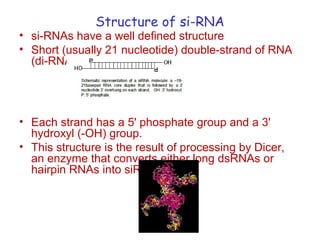
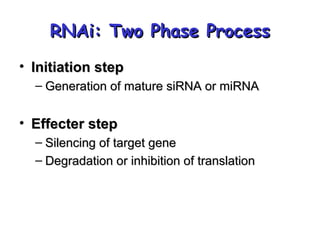
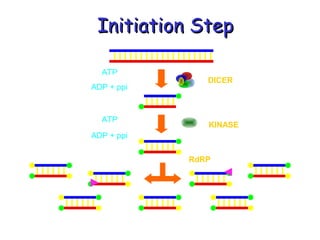
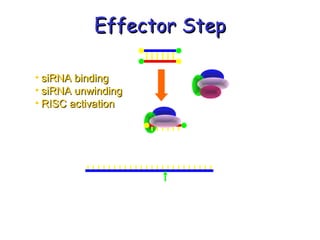
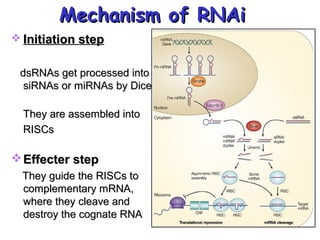
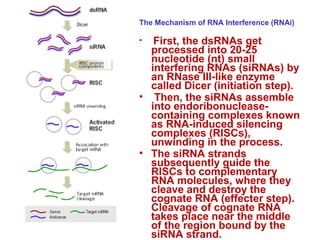
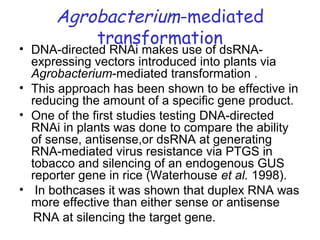
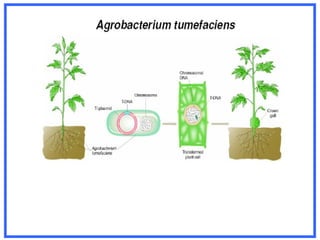
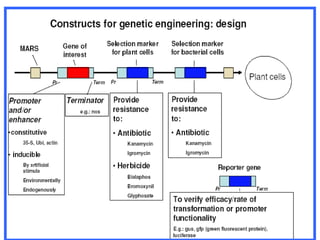
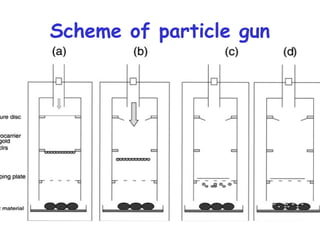
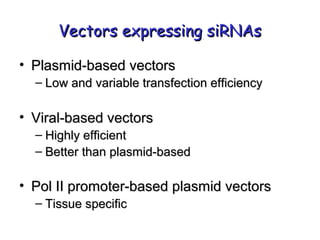
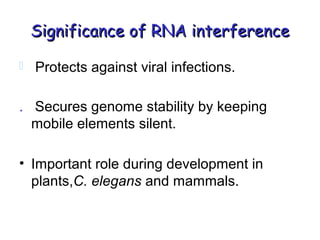
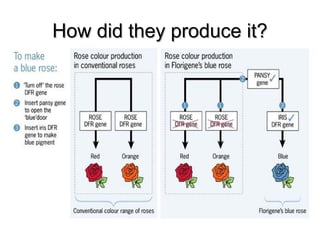
This document provides an overview of RNA interference (RNAi) and its applications in crop improvement. It discusses the history and discovery of RNAi, the mechanism of RNAi involving initiation by Dicer and effector function of RISC complexes, and methods of transforming plants with RNAi constructs. Applications of RNAi described in the document include modifying traits in rice, maize, barley, cotton, jute, tomato, lathyris, and coffee to improve nutritional quality, increase yields, confer virus resistance, and remove toxic compounds.