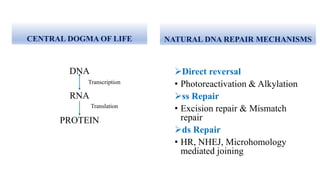
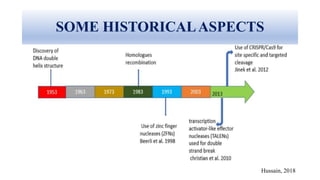
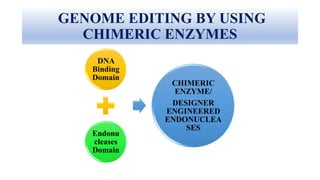
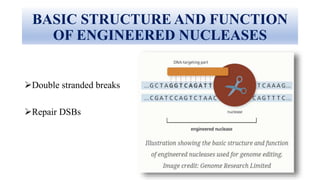
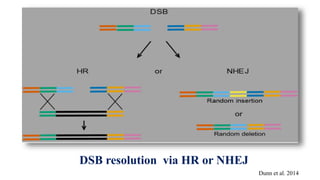
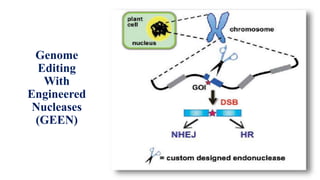
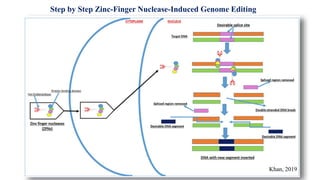
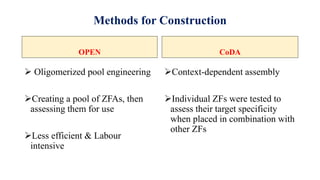
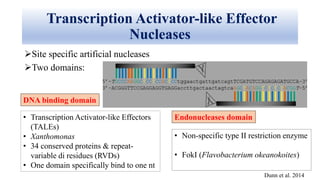
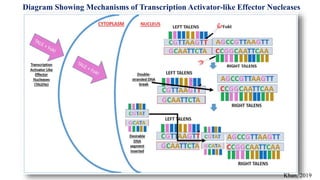
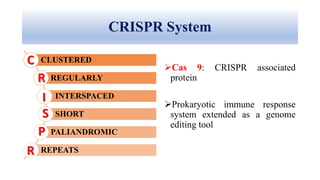
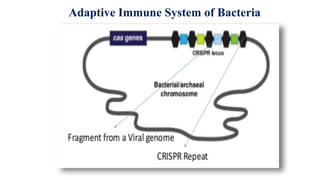
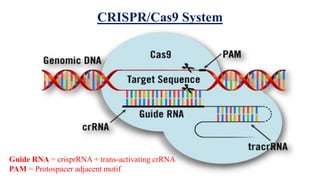
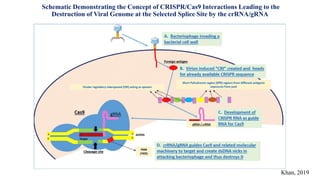
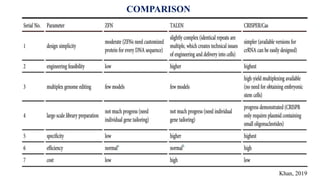
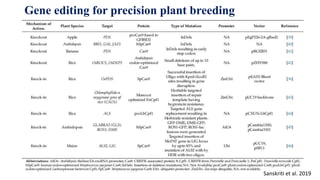
Genome editing allows specific changes to be made to the DNA of cells and organisms. Key techniques include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR-Cas9 system. ZFNs and TALENs use a DNA-binding domain fused to a nuclease to cut the DNA at a targeted location. CRISPR-Cas9 uses a guide RNA to target the Cas9 nuclease. Double-strand breaks caused by these nucleases can be repaired by non-homologous end joining or homology-directed repair, allowing genes to be knocked out, knocked down, or replaced. Genome editing holds promise for applications like disease