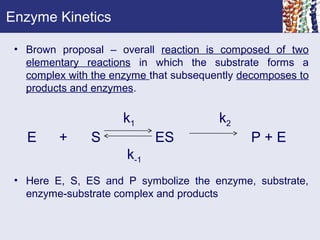
Enzyme kinetics is the study of reaction rates catalyzed by enzymes. Michaelis and Menten developed an equation to describe enzyme kinetics based on the assumptions that enzyme-substrate complexes form rapidly and at steady state. The Michaelis-Menten equation relates reaction rate to substrate concentration and can be linearized using double reciprocal plots. Enzyme kinetics are affected by factors like pH, temperature, and substrate concentration, and some enzymes use two substrates in either sequential or double displacement mechanisms.


![Enzyme Kinetics
• According to this model
• When the substrate concentration becomes high enough
to entirely convert the enzyme to the ES form, the
second step of the reaction becomes rate limiting step.
• The overall reaction rate becomes insensitive to further
increase in substrate concentration.
• The general expression of the velocity (rate) of this
reaction is
][
][
2
ESk
dt
Pd
v ==](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-4-320.jpg)
![Enzyme Kinetics
• The overall rate of production of [ES] – Difference between the
rates of elementary reactions leading to its appearance and
those resulting in its disappearance.
• At this point, an assumption is required to achieve an analytical
solution.
• The rapid equilibrium assumption
• Michaelis - Menten Approach.
• The steady-state assumption.
• Briggs and Haldane Approach.
][2][1]][[1
][
ESkESkSEk
dt
ESd
−−−=
EP
k
ES +→ 2
E+S
K-1
K1](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-5-320.jpg)
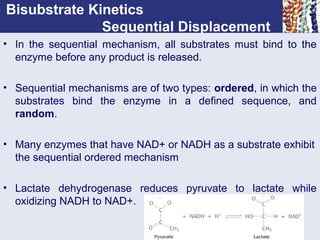
![Michaelis - Menten Approach
The rapid equilibrium assumption:
• Assumes a rapid equilibrium between the
enzyme and substrate to form an [ES] complex.
• The equilibrium constant Km can be expressed by
the following equation in a dilute system.
EP
k
ES +→ 2
E+S
K-1
K1
][1]][[1 ESkSEk −=
][
]][[
1
1
ES
SE
k
k
Km == −](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-6-320.jpg)
![Michaelis - Menten Approach
• Since the enzyme is not consumed, the
conservation equation on the enzyme yields
• Then rearrange the equilibrium constant
equation
• Substituting [E] in the above equation with
enzyme mass conservation equation
][]0[][ ESEE −=
][
]][[
1
1
ES
SE
k
k
Km == −
mK
SE
ES
]][[
][ ==
mK
SESE
ES
]])[[]([
][ 0 −
==](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-7-320.jpg)
![Michaelis - Menten Approach
mK
SESE
ES
]])[[]([
][ 0 −
==
]][[]][[][ 0 SESSEKES m −==
]][[]][[][ 0 SESESKES m ==+
]][[])[]([ 0 SESKES m ==+
][
]][[
][ 0
SK
SE
ES
m +
==](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-8-320.jpg)
![Michaelis - Menten Approach
• Then the rate of production formation v can
be expressed in terms of [S]
• Where
][
][
][
]][[
][
][ 02
2
SK
SV
SK
SEk
ESk
dt
Pd
v
mm +
=
+
=== max
][ 02
EkV =max](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-9-320.jpg)
![Steady State Assumption (SSA)
• Progress curve for the
components of a simple
michaelis-Menten
reaction
• Except the transition
phase of the reaction
(before shaded block)
[ES] remains constant
until the substrate is
nearly exhausted.
• Hence synthesis of ES
must equals to its
consumption over the
course of reaction i.e. ES
maintain steady state](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-10-320.jpg)
![•Now: Base on steady state assumption, d[ES]/dt = 0
•d[ES]/dt = k1[E][S] –k-1[ES] – k2[ES] = 0
(steady state assumption)
•solve for [ES] (do some algebra)
•[ES] = [E][S] k1/(k-1 + k2)
•Define KM (Michealis Constant)
•KM = (k-1 + k2)/k1 => [ES] = [E][S]/KM
SSA and Rate Equation](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-11-320.jpg)
![• Substitute in KM = [E][S]/[ES]][]0[][ ESEE −=
][
]])[[]([ 0
ES
SESE
Km
−
=
];])[[]([][ 0 SESEESKm −= ]][[]][[][ 0 SESSEKES m −==
]][[]][[][ 0 SESESKES m ==+
]][[])[]([ 0 SESKES m ==+
][
]][[
][ 0
SK
SE
ES
m +
==
SSA and Rate Equation](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-12-320.jpg)
![SSA lead to Michaelis - Menten
• Then the rate of production formation v can
be expressed in terms of [S]
• Where
• Michaelis Menten Equation
][
][
][
]][[
][
][ 02
2
SK
SV
SK
SEk
ESk
dt
Pd
v
mm +
=
+
=== max
][ 02
EkV =max
][
][
SK
SV
v
m +
= max](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-13-320.jpg)
![Michaelis Menten Equation
• Michaelis-Menten equation, the rate equation for
a one-substrate enzyme-catalyzed reaction.
• It is a statement of the quantitative relationship
between the initial velocity V0, the maximum velocity
Vmax, and the initial substrate concentration [S], all
related through the Michaelis constant Km.](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-14-320.jpg)
![Michaelis Menten Equation
• Numerical relationship emerges from the Michaelis-
Menten equation in the special case when V0 is
exactly one-half of Vmax
• On dividing by Vmax we obtained
• Solving for Km, we get Km + [S] = 2[S]
Km = [S] when
maxVv
2
1
0 =](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-15-320.jpg)

![Vmax
• Considering the total enzyme concentration the
maximal rate, that the enzyme can attain is Vmax,.
• Vmax is equal to the product of the catalytic rate
constant (kcat) and the concentration of the enzyme.
• The Michaelis-Menten equation can then be
rewritten as V= Kcat [Enzyme] [S] / (Km + [S]).
• Kcat is equal to K2, and it measures the number of
substrate molecules "turned over" by enzyme per
second.
• The higher the Kcat is, the more substrates get
turned over in one second.](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-17-320.jpg)
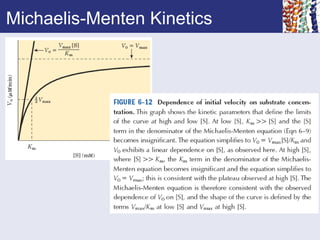
![Features of Michaelis-Menten
• Assumes the formation of Enzyme substrate
complex
• Assumes that the ES complex is in rapid equilibrium
with free enzyme
• Breakdown of ES to form products assumed to be
slower than
1. Formation of ES and
2. Breakdown of ES to reform E and S
][
][max
0
SK
SV
v
m +
=](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-19-320.jpg)
![Michaelis-Menten Kinetics
• KA is an equilibrium association constant (units: M-1
)
• KD is an equilibrium dissociation constant (units: M)
• Tight binding implies a low dissociation constant
and a high association constant
]][[
][
SE
ES
KA =
][
]][[
ES
SE
KD =](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-20-320.jpg)
![Transformations of the Michaelis-Menten
Equation: The Double-Reciprocal Plot
• The direct measurement of the numeric value of Vmax
and therefore the calculation of Km often requires
impractically high concentrations of substrate to
achieve saturating conditions
• The Michaelis-Menten equation
can be algebraically transformed
into equations that are more useful
in plotting experimental data.
][
][max
0
SK
SV
v
m +
=](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-21-320.jpg)
![Lineweaver-Burk Equation
• Starting with the MM equation
• Reciprocal of MM equation
• Lineweaver-Burk Equation
• Equation is the equation for a straight line, y = ax +
b, where y = 1/v0 and x = 1/[S].
][
][max
0
SK
SV
v
m +
=
maxmax0
1
][
1
VSV
K
v
m
+=
maxmax0
1
][
1
)(
1
VSV
K
v
m
+=](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-22-320.jpg)
![Lineweaver-Burk Equation
• A plot of 1/v0 as y as a function of 1/[S] as x therefore
gives a straight line whose y intercept is 1/Vmax and
whose slope is Km/Vmax.
• Such a plot is called a
double reciprocal or
Lineweaver-Burk plot
• Setting the y term of equation
equal to zero and solving for
x reveals that the x intercept
is −1/Km](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-23-320.jpg)
![Lineweaver-Burk Equation
• Lineweaver-Burk plot, has the great
advantage of allowing a more accurate
determination of Vmax, which can only be
approximated from a simple plot of V0 versus
[S]
• The double-reciprocal plot of enzyme reaction
rates is very useful in distinguishing between
certain types of enzymatic reaction
mechanisms.](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-24-320.jpg)
![Kinetics of Isosteric enzymes
• Isosteric enzymes
(with only one enzyme
conformation, 1), the
efficiency of
substrate binding
(dashed curve)
declines constantly
with increasing [A],
because the number
of free binding sites is
constantly decreasing.](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-25-320.jpg)
![Kinetics of allosteric enzymes
• Allosteric enzymes, the
binding efficiency initially
rises with increasing [A],
because the free enzyme
is present in a low-affinity
conformation (square
symbols), which is
gradually converted into a
higher-affinity form(round
symbols) as a result of
binding with A.
• It is only at high [A] values
that a lack of free binding
sites becomes noticeable
and the binding strength
decreases again.](https://image.slidesharecdn.com/enzymekinetics-160123162523/85/Enzyme-kinetics-26-320.jpg)