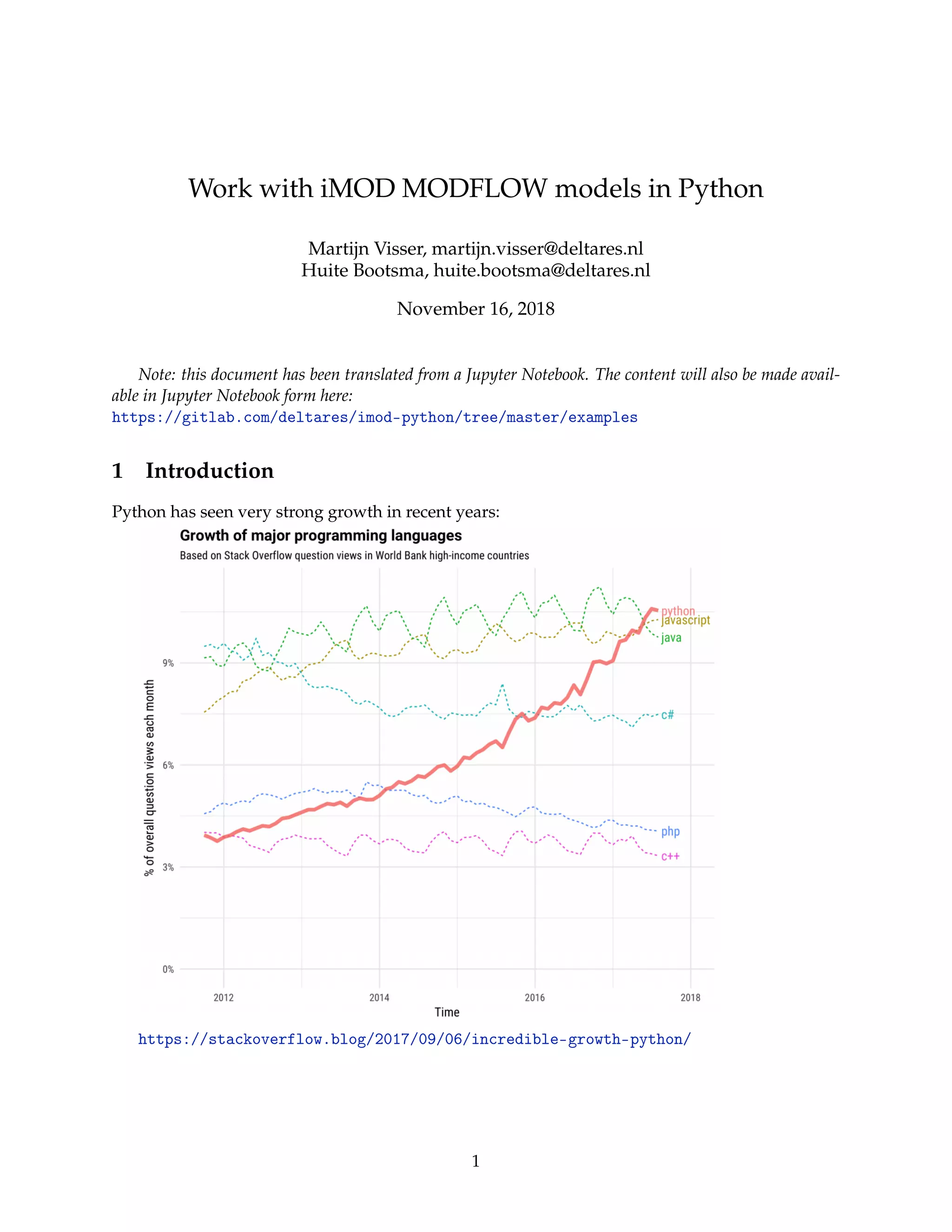
This document provides an overview of using the imod Python package for working with MODFLOW models, highlighting its installation and capabilities for technical computing with libraries like pandas and xarray. It includes a tutorial on converting Excel data to model inputs, building a groundwater flow model, and visualizing results. Key technical details and code snippets are presented throughout to facilitate understanding and implementation.

![1.2 What’s so great about pandas?
In [1]: import pandas
import matplotlib.pyplot as plt
%matplotlib inline
In [2]: fig = plt.figure(figsize=(25, 10))
df = pandas.read_csv("groundwater_timeseries.csv", index_col=1, parse_dates=[1])
df["head"].plot()
df["head"].rolling(window=180, center=True).mean().plot()
Out[2]: <matplotlib.axes._subplots.AxesSubplot at 0x8d20f60>
1.2.1 xarray versus numpy
# numpy style
>>> array[[0, 1, 3], :, :].max(axis=2)
# xarray style
>>> ds.sel(time="2017-11-28").max(dim="station")
4](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-4-320.jpg)
![2 Convert Excel to IPF
In [3]: import pandas
import imod
In [4]: df = pandas.read_excel("geophysical_measurements.xlsx")
In [6]: df.head()
Out[6]: CPT_name Depth_m Depth_m_NAP Sonic_velocity_m/sec Xcoord Ycoord
0 1210032_02 5.00 -3.74 1883.321 249255 607899
1 1210032_02 5.00 -3.74 1883.321 249255 607899
2 1210032_02 5.02 -3.76 1885.021 249255 607899
3 1210032_02 5.02 -3.76 1885.021 249255 607899
4 1210032_02 5.04 -3.78 1886.720 249255 607899
In [7]: df = df.rename(columns={"CPT_name": "id",
"Depth_m_NAP": "top",
"Xcoord": "x",
"Ycoord": "y"})
In [8]: imod.ipf.save("ipf_data/geophysical_measurements.ipf", df, itype="cpt")
5](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-5-320.jpg)
![In [9]: from collections import OrderedDict
import subprocess
import imod
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
%matplotlib inline
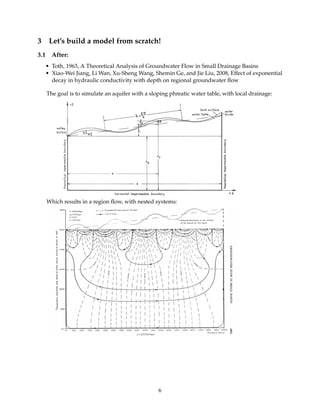
The phreatic water table is given by the following function (Toth, 1963):
zx(x) = z0 + x tan α + a
cos(α)
sin( bx
cos α )
Conductivity decreases exponentially with depth (Jiang et al., 2009):
k(z) = k0 exp[−A(zs − z)]
We can translate these functions into Python as follows:
In [10]: def phreatic_head(z0, x, a, alpha, b):
"""Synthetic ground surface, a la Toth 1963"""
return (z0 + x * np.tan(alpha) + a / np.cos(alpha)
* np.sin((b * x) / (np.cos(alpha))))
def conductivity(k0, z, A):
"""Exponentially decaying conductivity"""
return k0 * np.exp(-A * (1000.0 - z))
7](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-7-320.jpg)
![4 Generate the phreatic boundary condition
In [11]: z0 = 1000.0
x = np.arange(0.0, 6000.0, 10.0) + 5.0
a = 15.0
b = np.pi / 750.0
alpha = 0.02
In [12]: head_top = xr.DataArray(
data=phreatic_head(z0, x, a, alpha, b),
coords={"x": x},
dims=("x")
)
In [13]: fig = plt.figure(figsize=(10, 5))
head_top.plot()
Out[13]: [<matplotlib.lines.Line2D at 0xbc34d68>]
8](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-8-320.jpg)
![5 Generate conductivity
In [14]: ncol = x.size
nrow = 1
nlay = 100
coords = {"layer": np.arange(1, 101), "y": [5.0], "x": x}
dims = ("layer", "y", "x")
bnd = xr.DataArray(
data=np.full((nlay, nrow, ncol), 1.0),
coords=coords,
dims=dims,
)
In [15]: k0 = 1.0 # m/d
A = 0.001
z = np.arange(0.0, 1000.0, 10.0) + 5.0
k_z = xr.DataArray(
data=conductivity(k0, z, A),
coords={"z": z},
dims=("z"),
)
k_z.coords["z"] = bnd.coords["layer"].values[::-1]
k_z = k_z.rename({"z": "layer"})
kh = bnd.copy() * k_z
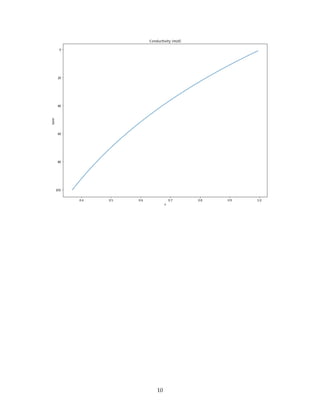
In [16]: fig = plt.figure(figsize=(13, 10))
k_z.plot(y="layer")
plt.title("Conductivity (m/d)")
plt.gca().invert_yaxis()
plt.xlabel("x")
Out[16]: Text(0.5,0,'x')
9](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-9-320.jpg)
![6 Generate the iMODFLOW model files
In [17]: model = OrderedDict()
model["bnd"] = bnd
# constant head in the first layer
model["bnd"].sel(layer=1)[...] = -1.0
model["kdw"] = kh * 10.0
model["vcw"] = 10.0 / kh
model["shd"] = bnd * head_top
# We have to an additional package for it to run...
model["rch"] = xr.full_like(bnd.sel(layer=1), 0.0)
In [18]: imod.write("toth", model)
Now let’s inspect these files, and run the model.
11](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-11-320.jpg)
![6.1 Load the results
In [19]: head = imod.idf.load("results/head/*.idf")
head
Out[19]: <xarray.DataArray 'head' (layer: 100, y: 1, x: 600)>
dask.array<shape=(100, 1, 600), dtype=float32, chunksize=(1, 1, 600)>
Coordinates:
* y (y) float64 5.0
* x (x) float64 5.0 15.0 25.0 35.0 ... 5.975e+03 5.985e+03 5.995e+03
* layer (layer) int32 1 2 3 4 5 6 7 8 9 10 ... 92 93 94 95 96 97 98 99 100
Attributes:
res: (10.0, 10.0)
transform: (10.0, 0.0, 0.0, 0.0, -10.0, 10.0)
In [20]: fig = plt.figure(figsize=(10, 6))
head.isel(y=0).plot()
plt.gca().invert_yaxis()
12](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-12-320.jpg)
![7 Look at streamlines
In [21]: vz = imod.idf.load("results/bdgflf/*.idf").isel(y=0).values[:]
vx = imod.idf.load("results/bdgfrf/*.idf").isel(y=0).values[:]
In [22]: f, (ax1, ax2) = plt.subplots(ncols=1, nrows=2, sharex=True,
gridspec_kw={'height_ratios': [0.2, 0.8]},
figsize=(15, 5)
)
ax1.plot(head_top["x"], head_top.values, color="k")
ax1.set_ylabel("head(m)", size=16)
ax2.streamplot(x, z[:], vx, vz, arrowsize=2)
ax2.invert_yaxis()
ax2.set_ylabel("layer", size=16)
ax2.set_xlabel("x", size=16)
f.set_figheight(10.0)
13](https://image.slidesharecdn.com/11-181128110131/85/DSD-INT-2018-Work-with-iMOD-MODFLOW-models-in-Python-Visser-Bootsma-13-320.jpg)