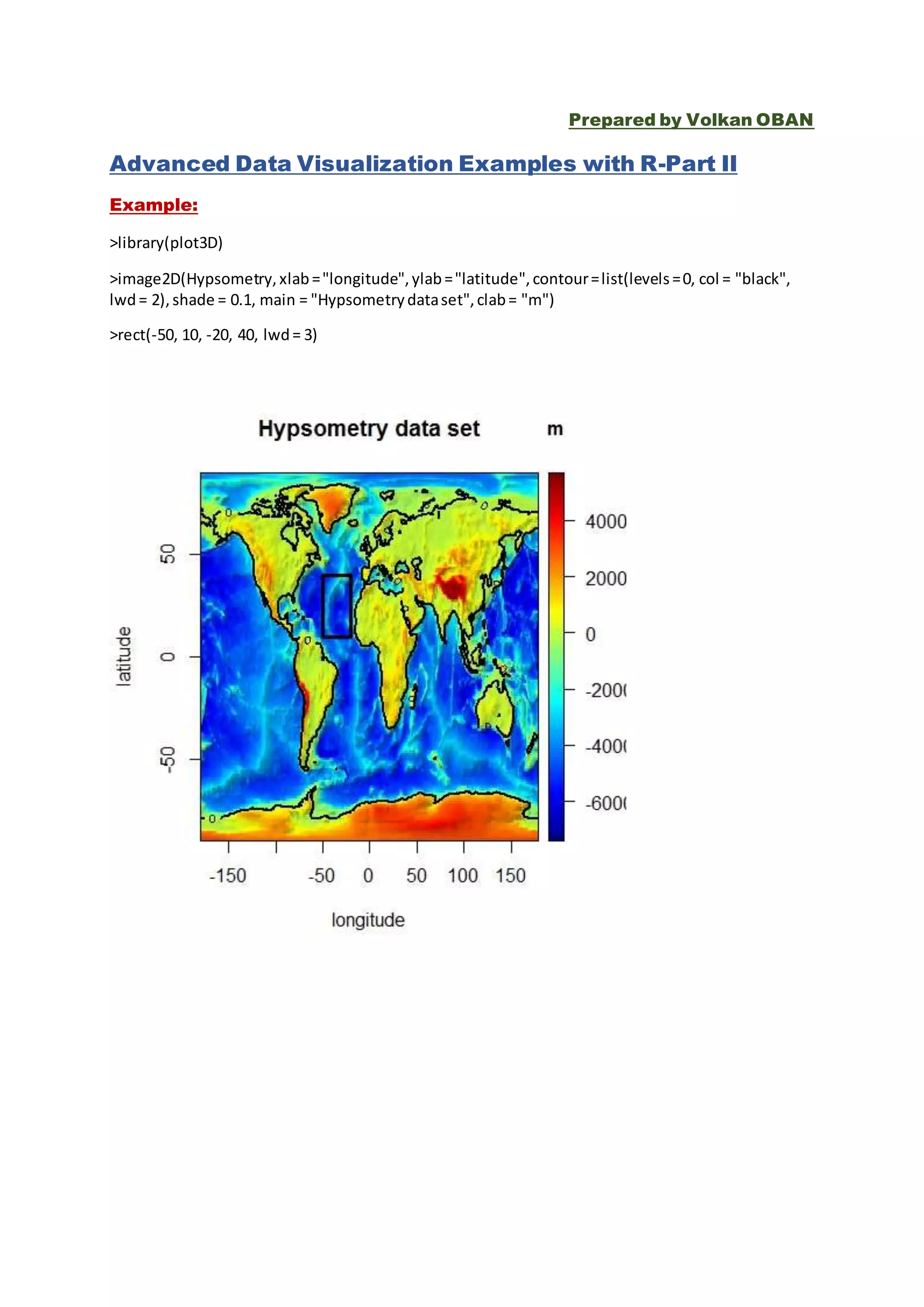
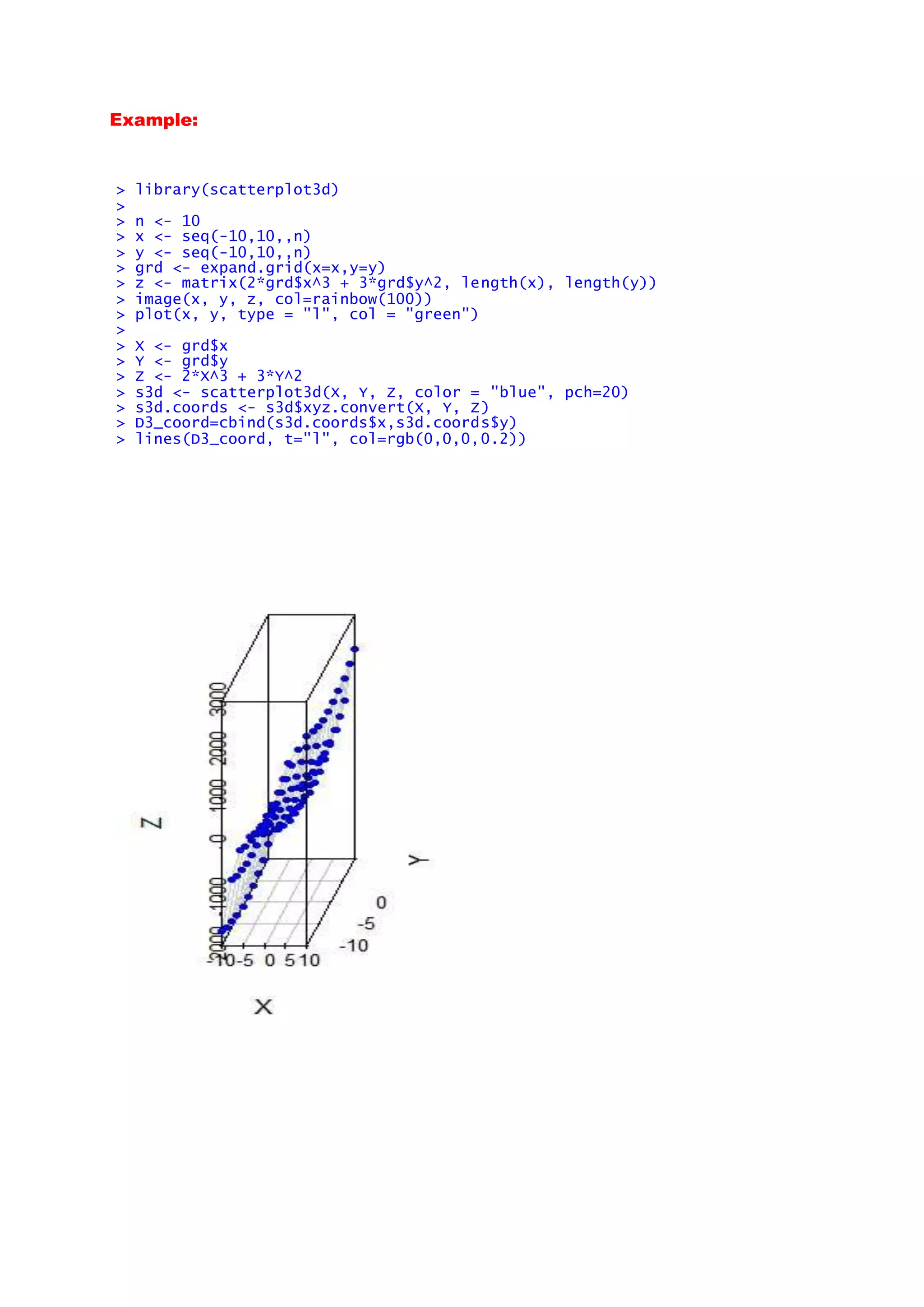
This document provides several examples of advanced data visualization techniques using R. It includes examples of 3D surface plots, contour plots, scatter plots and network graphs using various R packages like plot3D, scatterplot3D, ggplot2, qgraph and ggtree. Functions used include surf3D, contour3D, arrows3D, persp3D, image3D, scatter3D, qgraph, geom_point, geom_violin and ggtree. The examples demonstrate different visualization approaches for multivariate, spatial and network data.


![Example:
> persp3D(z = volcano, zlim = c(-60, 200), phi = 20,
colkey = list(length = 0.2, width = 0.4, shift = 0.15,
cex.axis = 0.8, cex.clab = 0.85), lighting = TRUE, lphi = 90,clab = c("","
height","m"), bty = "f", plot = FALSE)
> # create gradient in x-direction
> Vx <- volcano[-1, ] - volcano[-nrow(volcano), ]
> # add as image with own color key, at bottom
> image3D(z = -60, colvar = Vx/10, add = TRUE,colkey = list(length = 0.2,
width = 0.4, shift = -0.15,cex.axis = 0.8, cex.clab = 0.85),clab = c("","g
radient","m/m"), plot = FALSE)
> # add contour
> contour3D(z = -60+0.01, colvar = Vx/10, add = TRUE,col = "black", plot =
TRUE)](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-4-2048.jpg)

![Example:
require(plot3D)
lon <- seq(165.5, 188.5, length.out = 30)
lat <- seq(-38.5, -10, length.out = 30)
xy <- table(cut(quakes$long, lon),
cut(quakes$lat, lat))
xmid <- 0.5*(lon[-1] + lon[-length(lon)])
ymid <- 0.5*(lat[-1] + lat[-length(lat)])
par (mar = par("mar") + c(0, 0, 0, 2))
hist3D(x = xmid, y = ymid, z = xy,
zlim = c(-20, 40), main = "Earth quakes",
ylab = "latitude", xlab = "longitude",
zlab = "counts", bty= "g", phi = 5, theta = 25,
shade = 0.2, col = "white", border = "black",
d = 1, ticktype = "detailed")
with (quakes, scatter3D(x = long, y = lat,
z = rep(-20, length.out = length(long)),
colvar = quakes$depth, col = gg.col(100),
add = TRUE, pch = 18, clab = c("depth", "m"),
colkey = list(length = 0.5, width = 0.5,
dist = 0.05, cex.axis = 0.8, cex.clab = 0.8)))](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-7-2048.jpg)
![Example:
> library(maps)
> coplot(lat ~ long | depth, data = quakes, number=4,
panel=function(x, y, ...) {
usr <- par("usr")
rect(usr[1], usr[3], usr[2], usr[4], col="white")
map("world2", regions=c("New Zealand", "Fiji"),
add=TRUE, lwd=0.1, fill=TRUE, col="grey")
text(180, -13, "Fiji", adj=1, cex=0.7)
text(170, -35, "NZ", cex=0.7)
points(x, y, pch=".") })](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-8-2048.jpg)



![Example:
> library(AnalyzeFMRI)
Zorunlu paket yükleniyor: tcltk
Zorunlu paket yükleniyor: R.matlab
R.matlab v3.6.0 (2016-07-05) successfully loaded. See ?R.matlab for help
> a <- f.read.analyze.volume(system.file("example.img", package="AnalyzeFMR
I"))
> a <- a[,,,1]
> contour3d(a, 1:64, 1:64, 1.5*(1:21), lev=c(3000, 8000, 10000),
+ alpha = c(0.2, 0.5, 1), color = c("white", "red", "green"))
> # alternative masking out a corner
> m <- array(TRUE, dim(a))
> m[1:30,1:30,1:10] <- FALSE
> contour3d(a, 1:64, 1:64, 1.5*(1:21), lev=c(3000, 8000, 10000),
+ mask = m, color = c("white", "red", "green"))
> contour3d(a, 1:64, 1:64, 1.5*(1:21), lev=c(3000, 8000, 10000),
+ color = c("white", "red", "green"),
+ color2 = c("gray", "red", "green"),
+ mask = m, engine="standard",
+ scale = FALSE, screen=list(z = 60, x = -120))](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-12-2048.jpg)
![Example:
> nmix3 <- function(x, y, z, m, s) {
0.3*dnorm(x, -m, s) * dnorm(y, -m, s) * dnorm(z, -m, s) +
0.3*dnorm(x, -2*m, s) * dnorm(y, -2*m, s) * dnorm(z, -2*m, s) +
0.4*dnorm(x, -3*m, s) * dnorm(y, -3 * m, s) * dnorm(z, -3*m, s) }
> f <- function(x,y,z) nmix3(x,y,z,0.5,.1)
> n <- 20
> x <- y <- z <- seq(-2, 2, len=n)
> contour3dObj <- contour3d(f, 0.35, x, y, z, draw=FALSE, separate=TRUE)
> for(i in 1:length(contour3dObj))
contour3dObj[[i]]$color <- rainbow(length(contour3dObj))[i]
> drawScene.rgl(contour3dObj)
>](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-13-2048.jpg)



![Example:
library(plyr)
mean.prop.sw <- c(0.7, 0.6, 0.67, 0.5, 0.45, 0.48, 0.41, 0.34, 0.5, 0.33)
sd.prop.sw <- c(0.3, 0.4, 0.2, 0.35, 0.28, 0.31, 0.29, 0.26, 0.21, 0.23)
N <- 100
b <- barplot(mean.prop.sw, las = 1, xlab = " ", ylab = " ", col = "grey", c
ex.lab = 1.7,
cex.main = 1.5, axes = FALSE, ylim = c(0, 1))
axis(1, c(0.8, 2, 3.2, 4.4, 5.6, 6.8, 8, 9.2, 10.4, 11.6), 1:10, cex.axis =
1.3)
axis(2, seq(0, 0.8, by = 0.2), cex.axis = 1.3, las = 1)
mtext("Block", side = 1, line = 2.5, cex = 1.5, font = 2)
mtext("Proportion of Switches", side = 2, line = 3, cex = 1.5, font = 2)
l_ply(seq_along(b), function(x) arrows(x0 = b[x], y0 = mean.prop.sw[x], x1
= b[x],
y1 = mean.prop.sw[x] + 1.96 * sd.prop.sw[x]/sqrt(N), code = 2, length =
0.1,
angle = 90, lwd = 1.5))](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-17-2048.jpg)

![Example:
>library("psych")
> library("qgraph")
>
> # Load BFI data:
> data(bfi)
> bfi <- bfi[, 1:25]
>
> # Groups and names object (not needed really, but make the plots easier t
o
> # interpret):
> Names <- scan("http://sachaepskamp.com/files/BFIitems.txt", what = "chara
cter", sep = "n")
Read 25 items
>
> # Create groups object:
> Groups <- rep(c("A", "C", "E", "N", "O"), each = 5)
>
> # Compute correlations:
> cor_bfi <- cor_auto(bfi)
Variables detected as ordinal: A1; A2; A3; A4; A5; C1; C2; C3; C4; C5; E1;
E2; E3; E4; E5; N1; N2; N3; N4; N5; O1; O2; O3; O4; O5
>
> # Plot correlation network:
> graph_cor <- qgraph(cor_bfi, layout = "spring", nodeNames = Names, groups
= Groups, legend.cex = 0.6,
+ DoNotPlot = TRUE)
>
> # Plot partial correlation network:
> graph_pcor <- qgraph(cor_bfi, graph = "concentration", layout = "spring",
nodeNames = Names,
+ groups = Groups, legend.cex = 0.6, DoNotPlot = TRUE)
>
> # Plot glasso network:
> graph_glas <- qgraph(cor_bfi, graph = "glasso", sampleSize = nrow(bfi), l
ayout = "spring",
+ nodeNames = Names, legend.cex = 0.6, groups = Groups
, legend.cex = 0.7, GLratio = 2)](https://image.slidesharecdn.com/plot3d-160926212257/75/Advanced-Data-Visualization-Examples-with-R-Part-II-19-2048.jpg)






