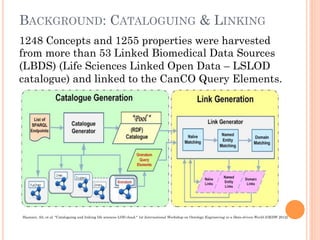
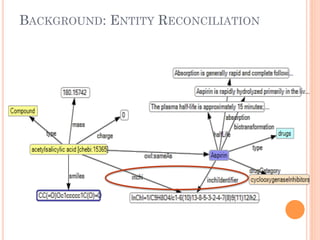
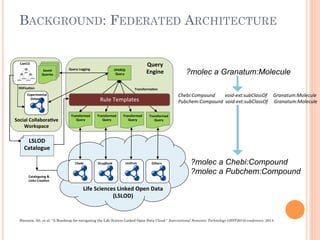
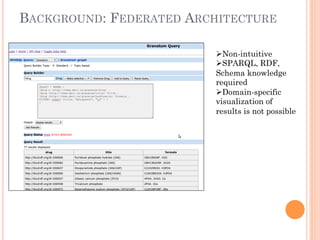
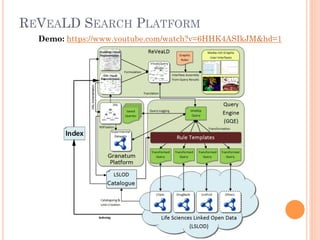
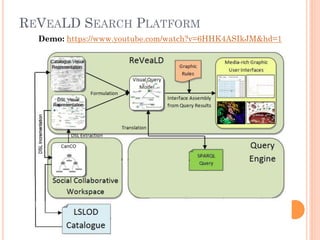
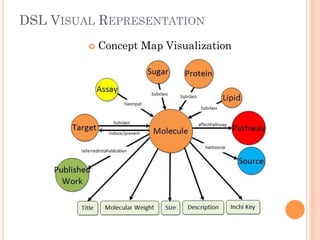
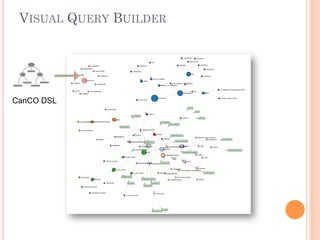
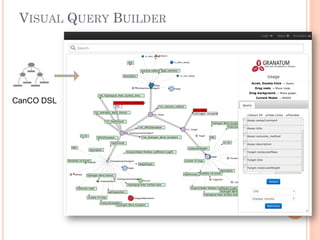
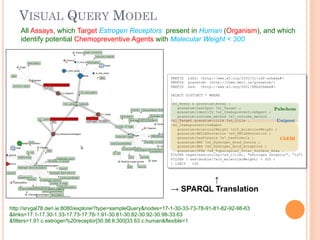
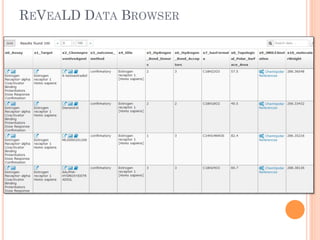
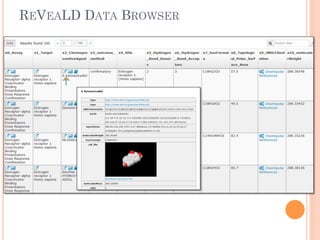
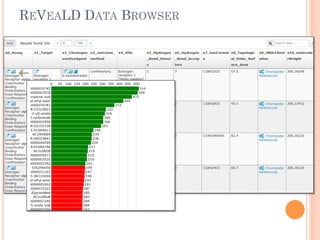
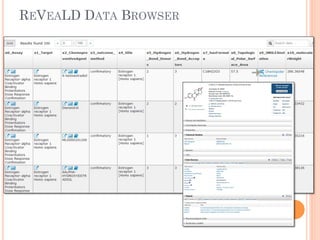
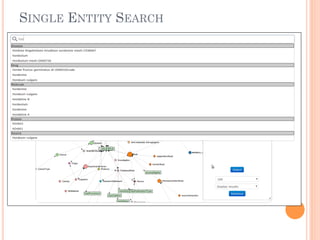
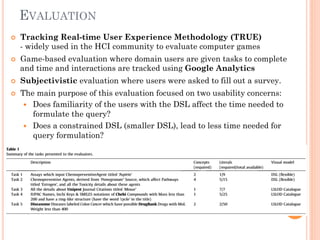
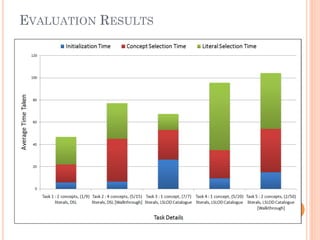
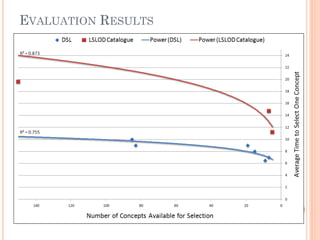
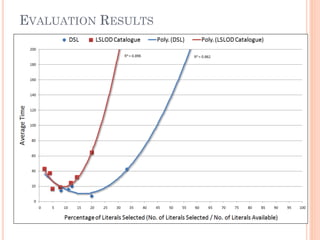
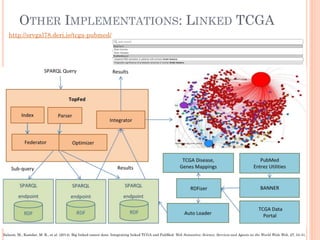
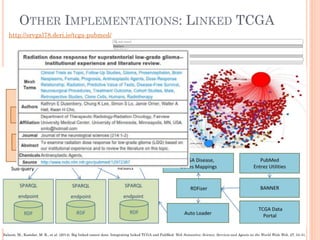
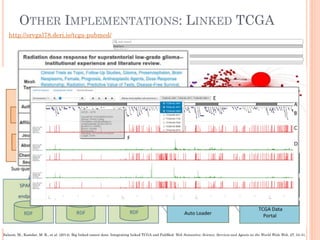
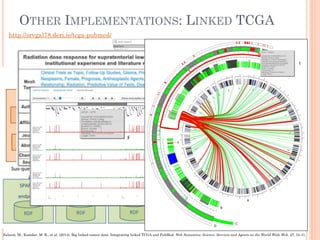
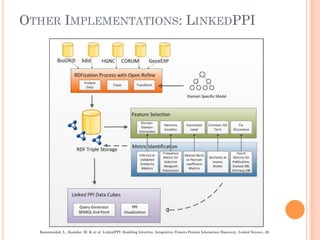
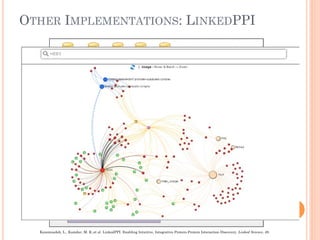
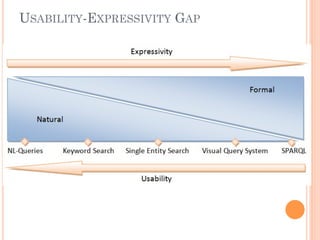
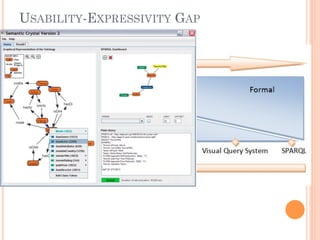
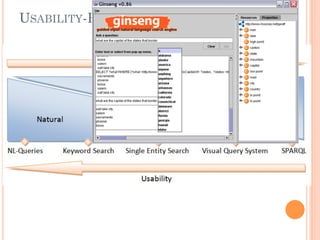
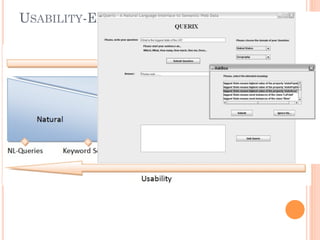
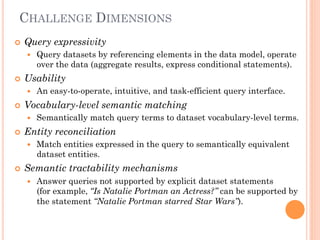
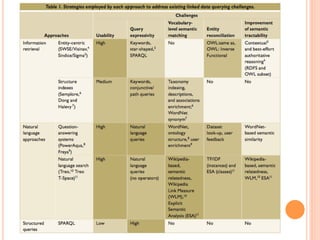
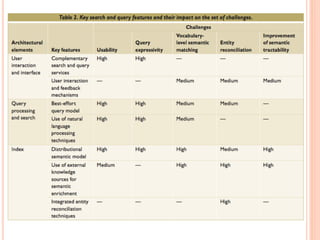
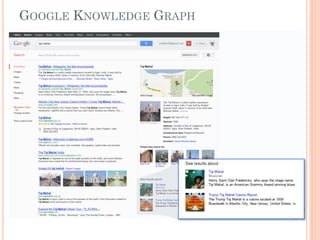
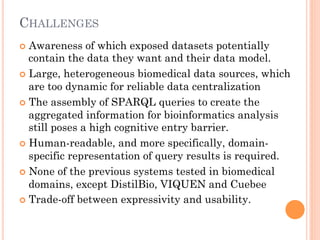
The document discusses advancements aimed at improving usability and expressivity in biomedical semantic search, particularly through user-driven platforms like Reveald, which enable intuitive query formulation and visualization of linked data. It highlights challenges including the complexity of querying heterogeneous data, a need for human-readable results, and the trade-off between user-friendliness and query expressivity. Future work includes enhancing domain-specific languages and incorporating user-generated elements to facilitate better data access in biomedical research.





![LIFE SCIENCES LINKED OPEN DATA CLOUD
~3 Billion Triples Life
Sciences
53 datasets
Cyganiak,R. and Jentzsch,A. (2014) The Linking Open Data cloud diagram. http://lod-cloud.net/ [Accessed: March 23, 2013]](https://image.slidesharecdn.com/noanimprotegemeetingpresentation2015-150410191304-conversion-gate01/85/Current-advances-to-bridge-the-usability-expressivity-gap-in-biomedical-semantic-search-and-visualizing-linked-data-21-320.jpg)