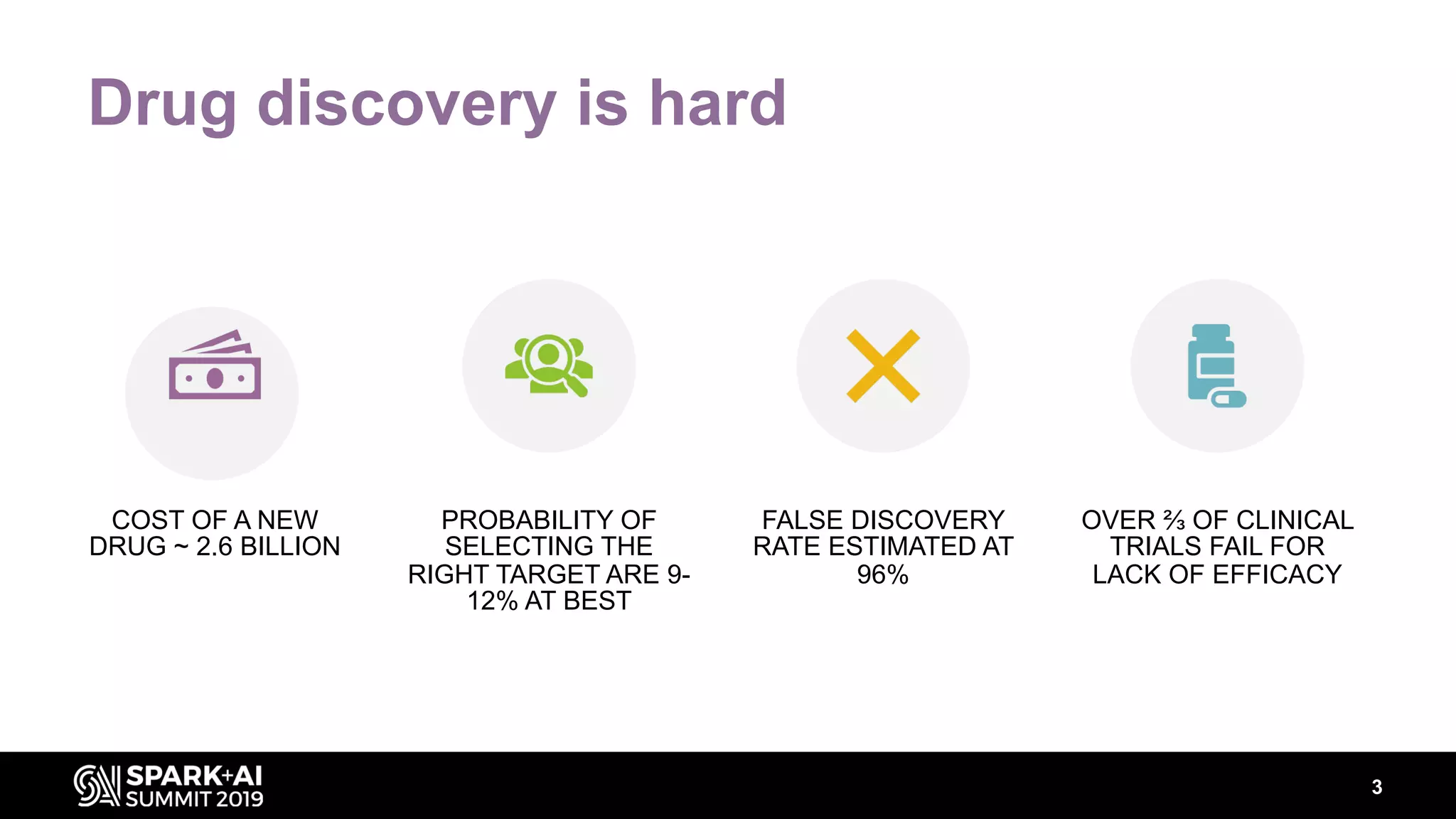
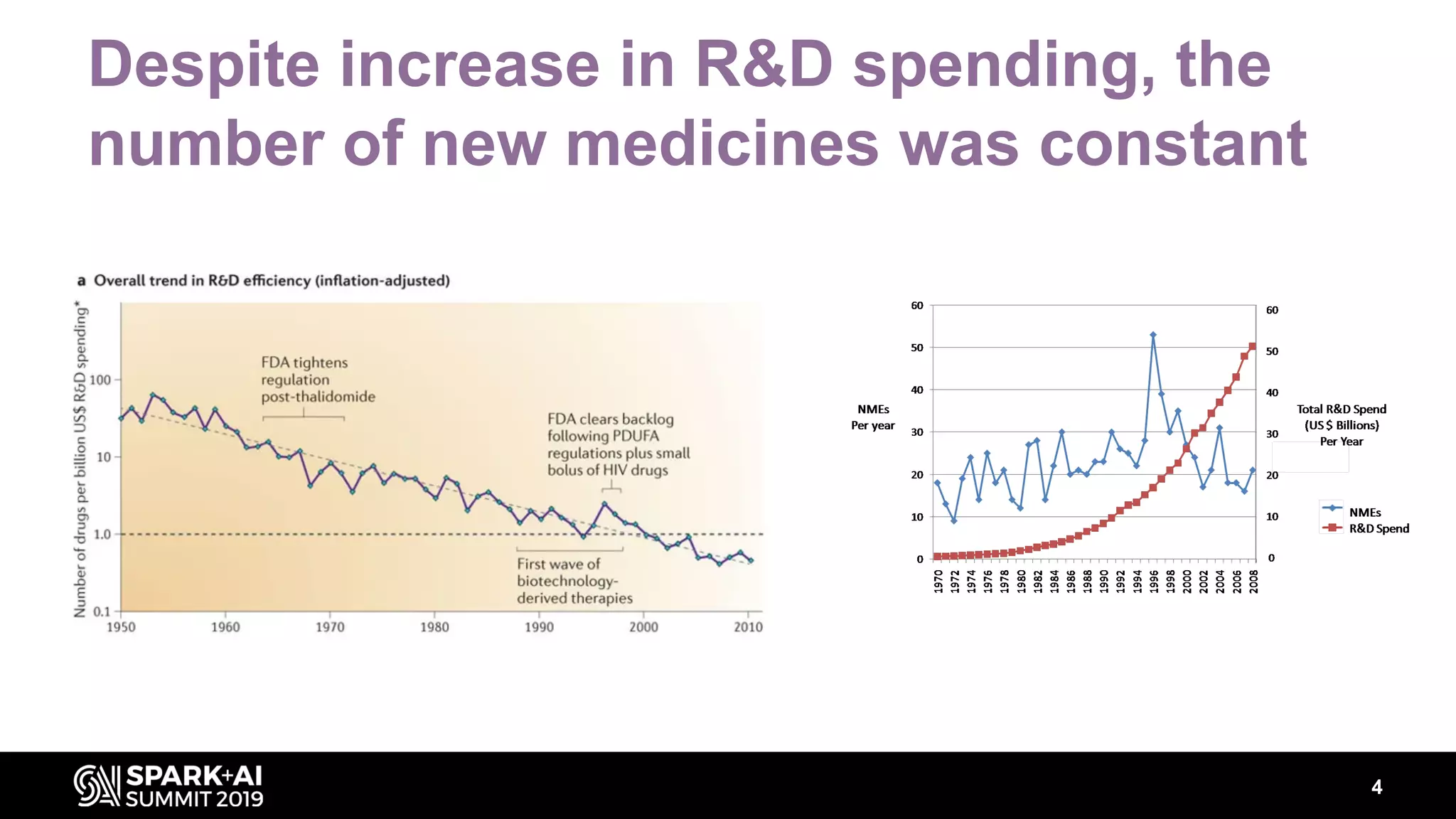
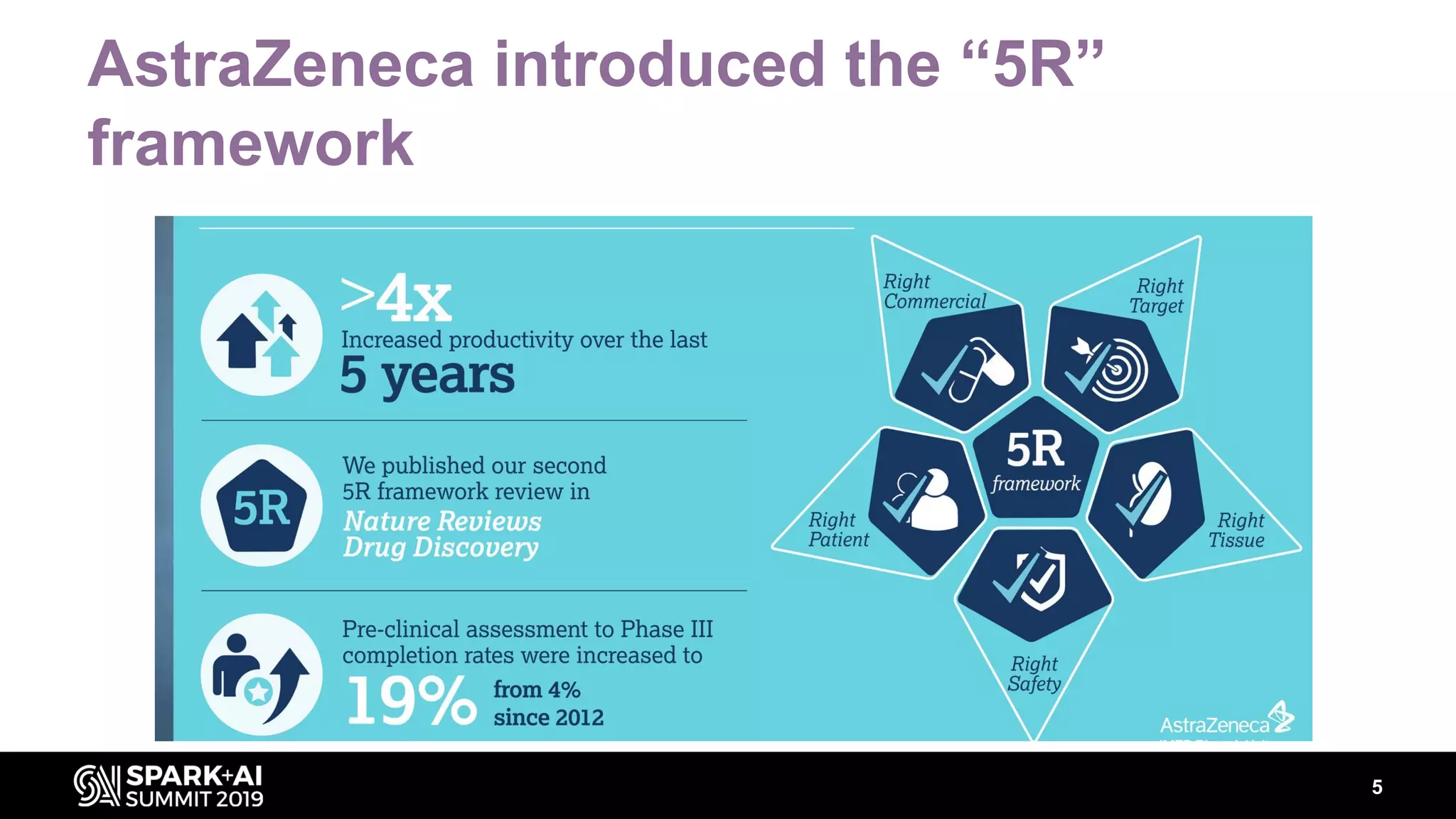
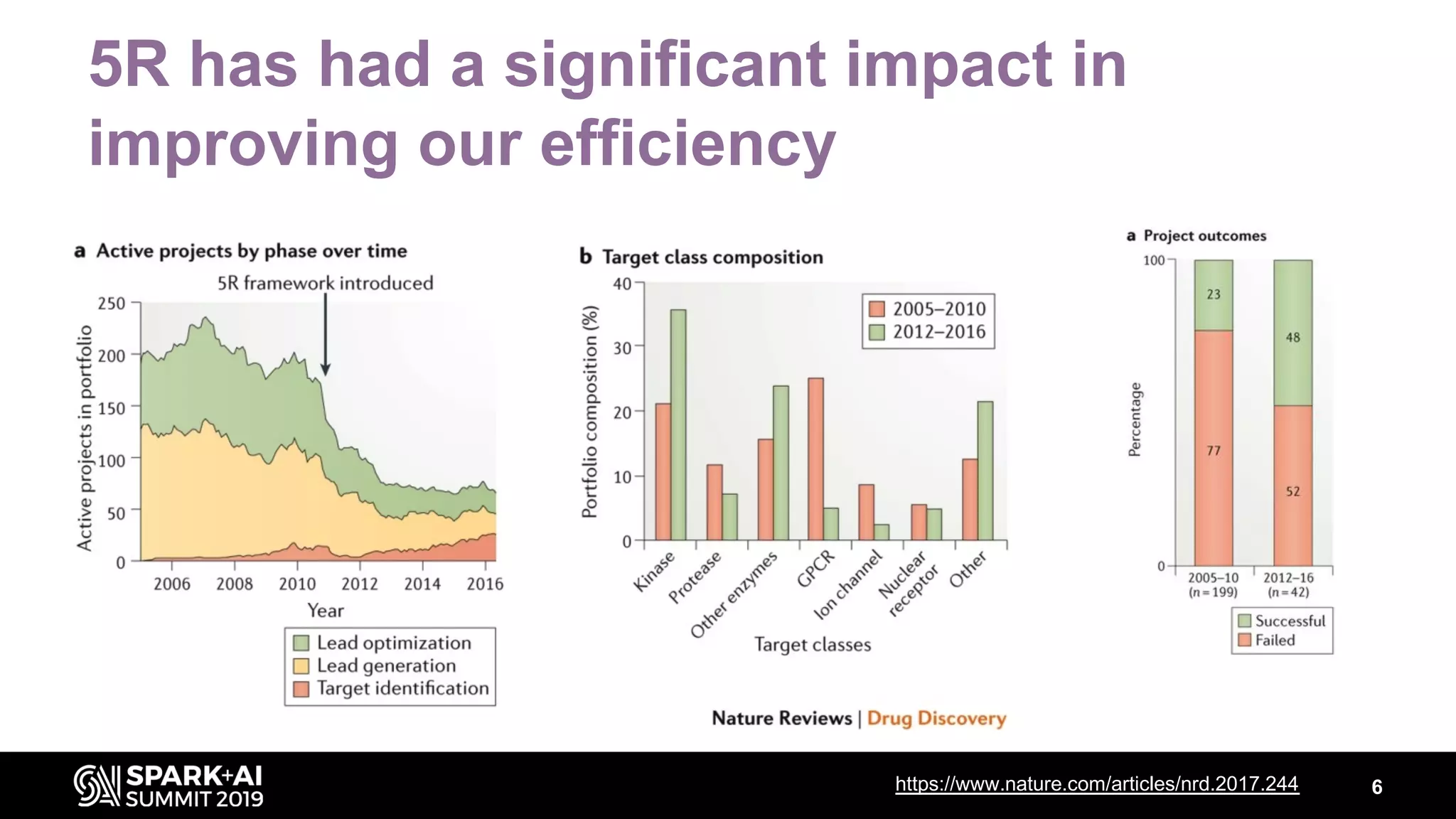
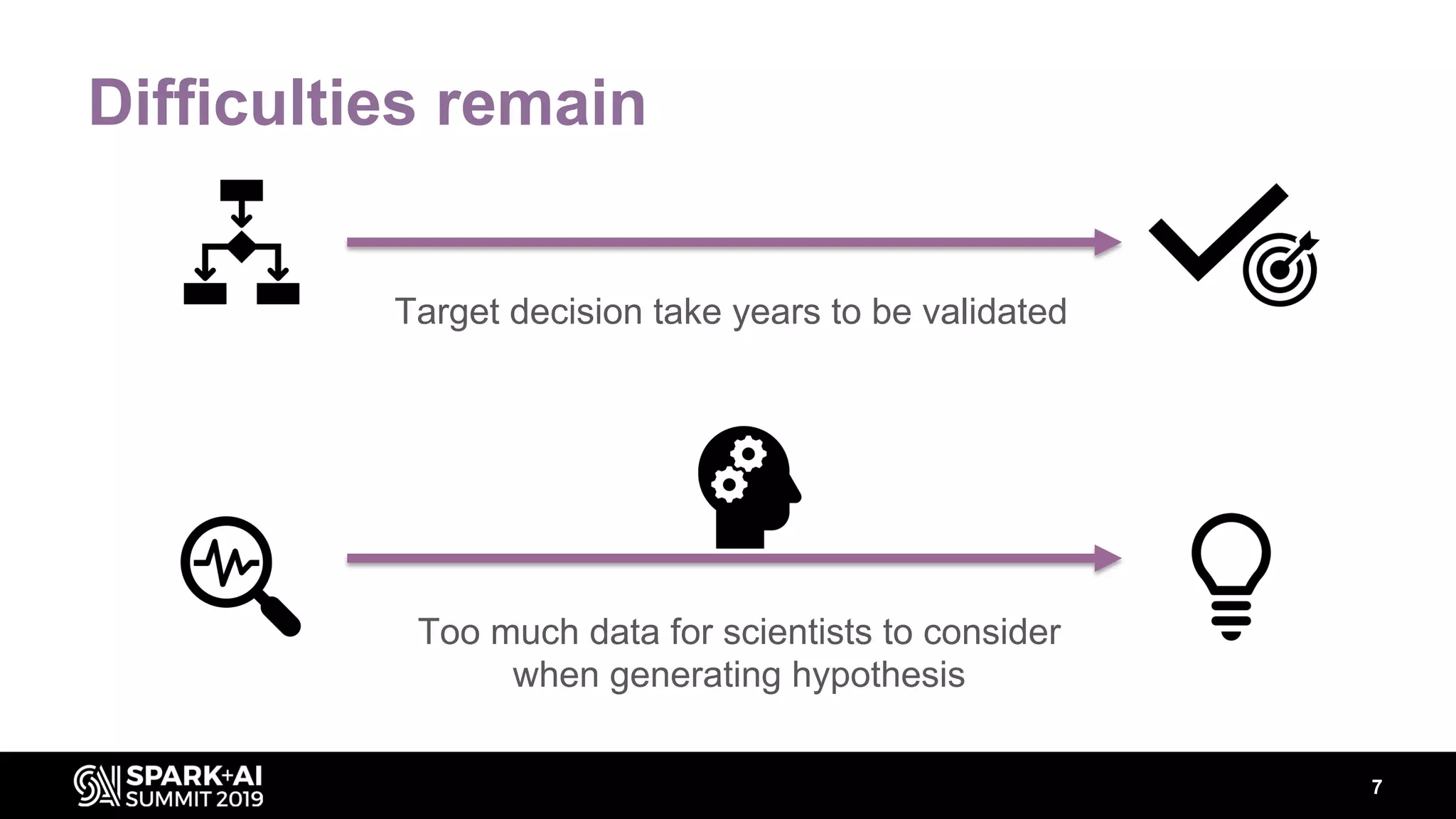
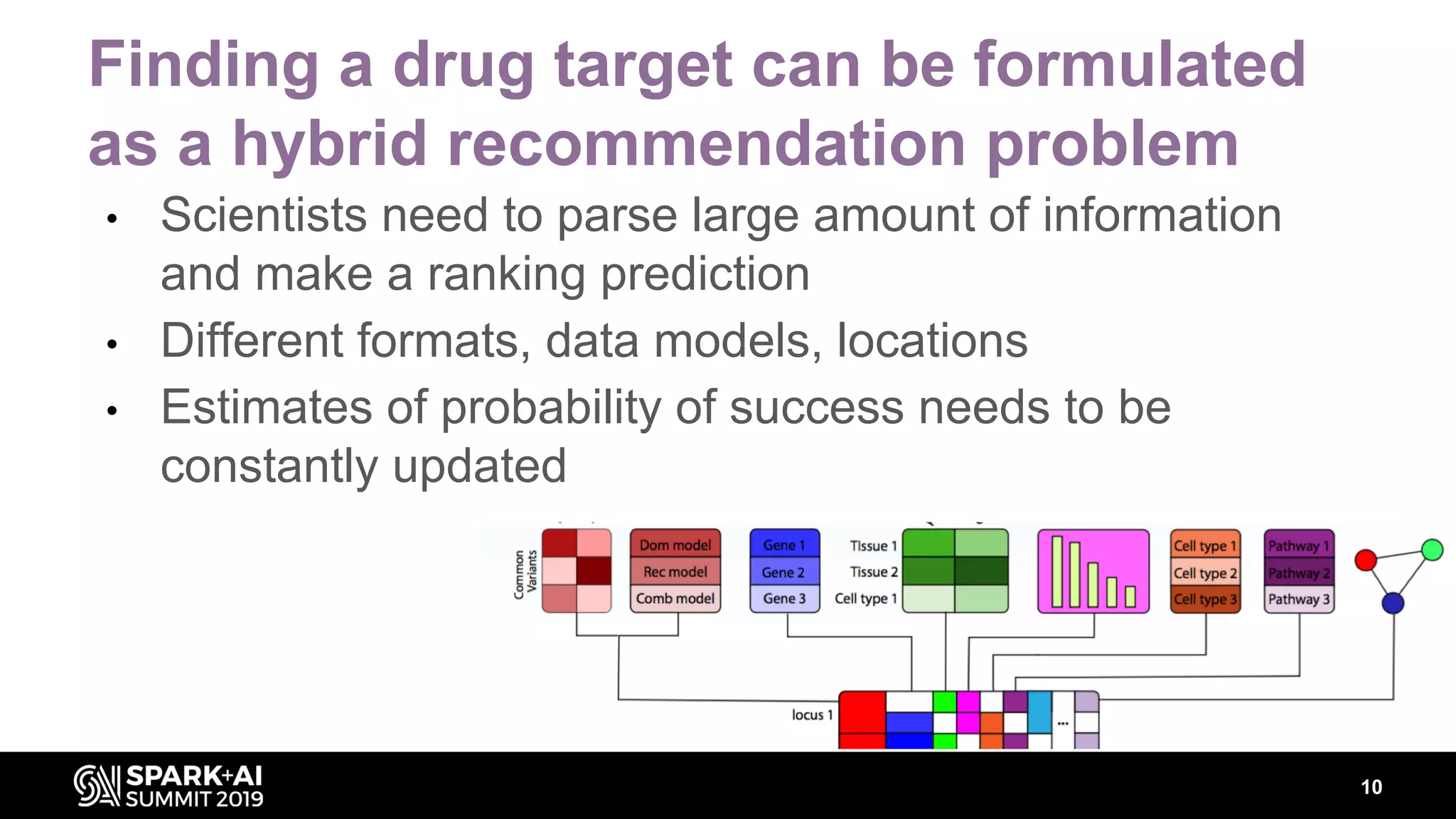
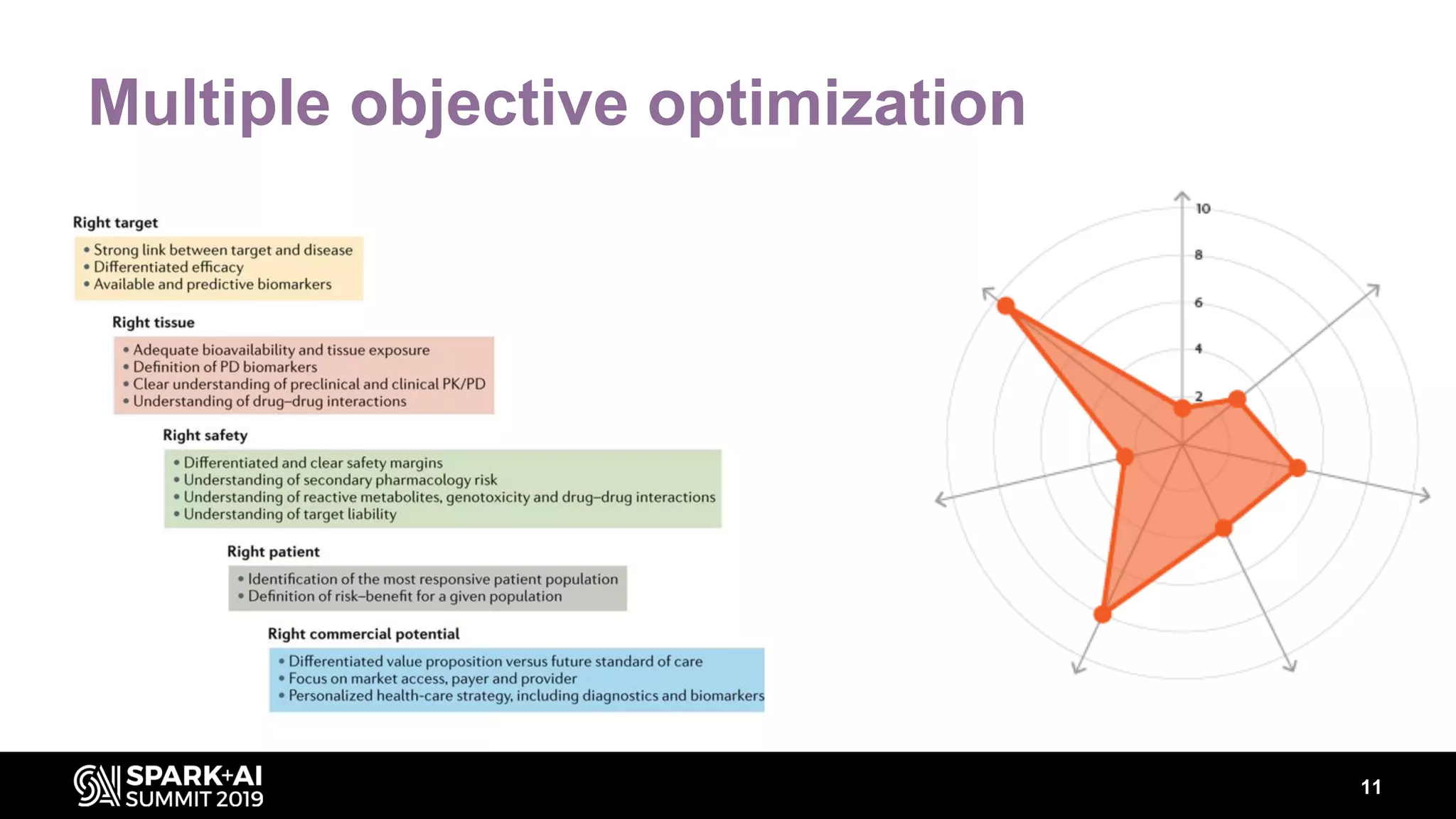
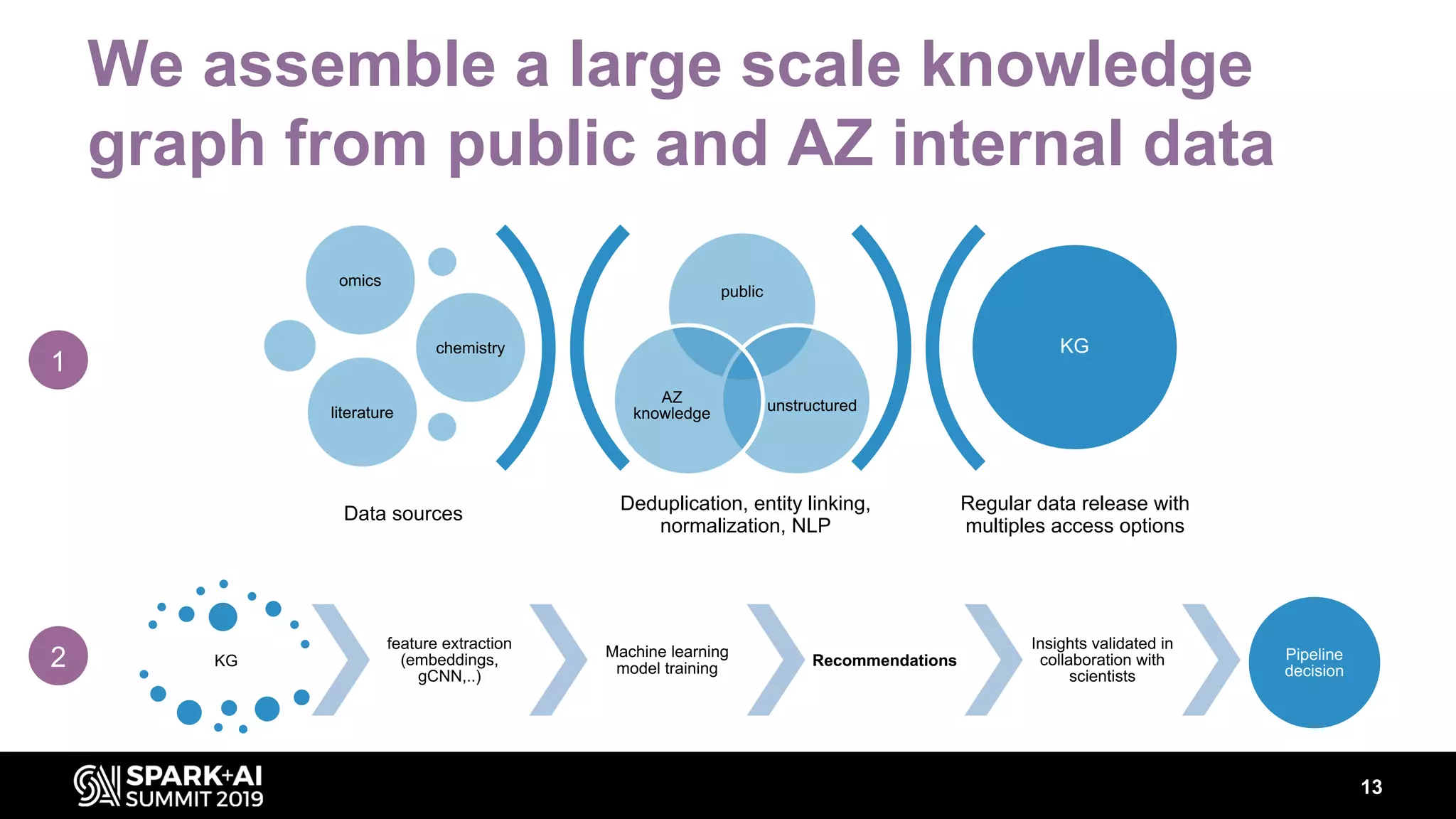
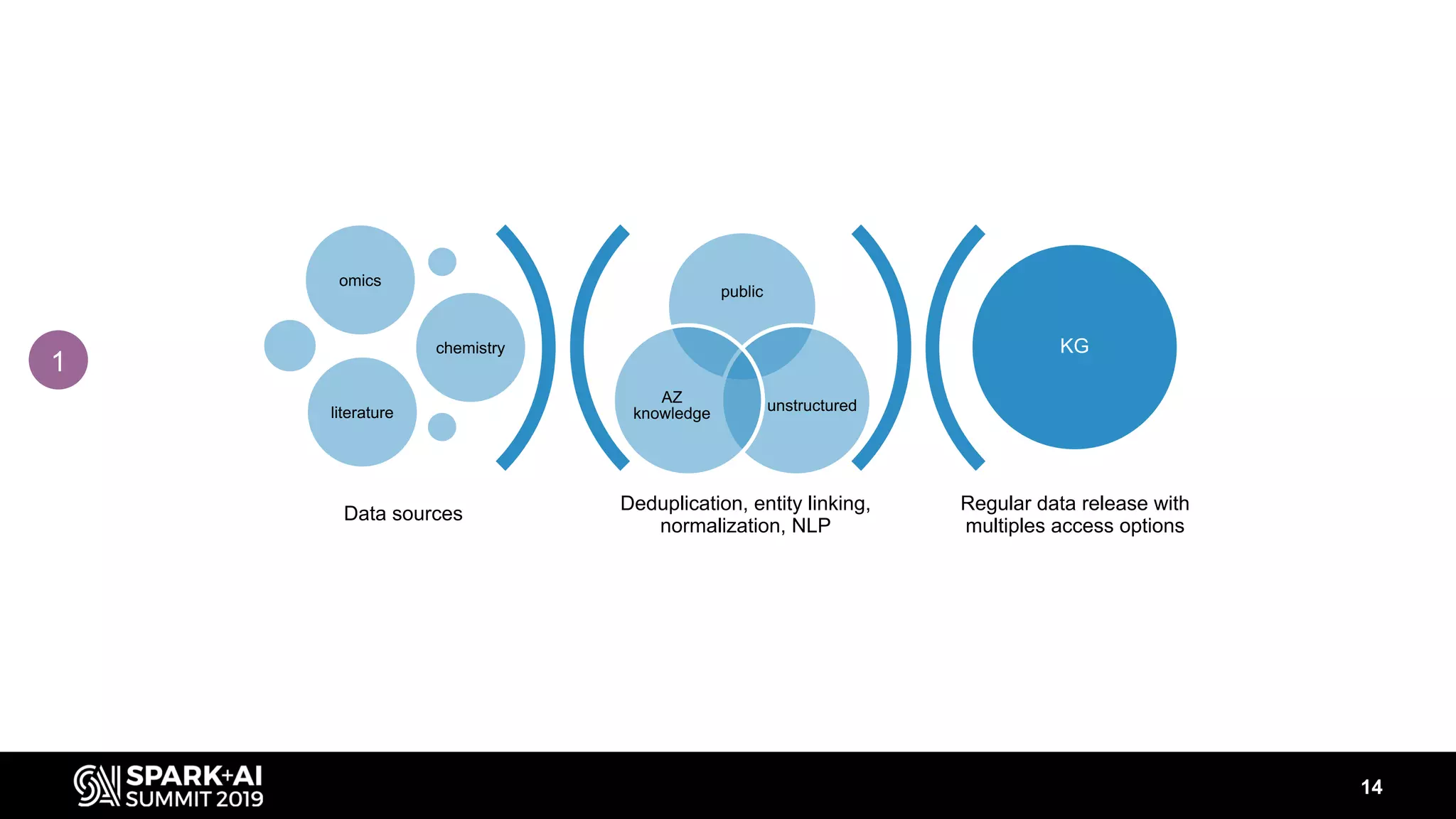
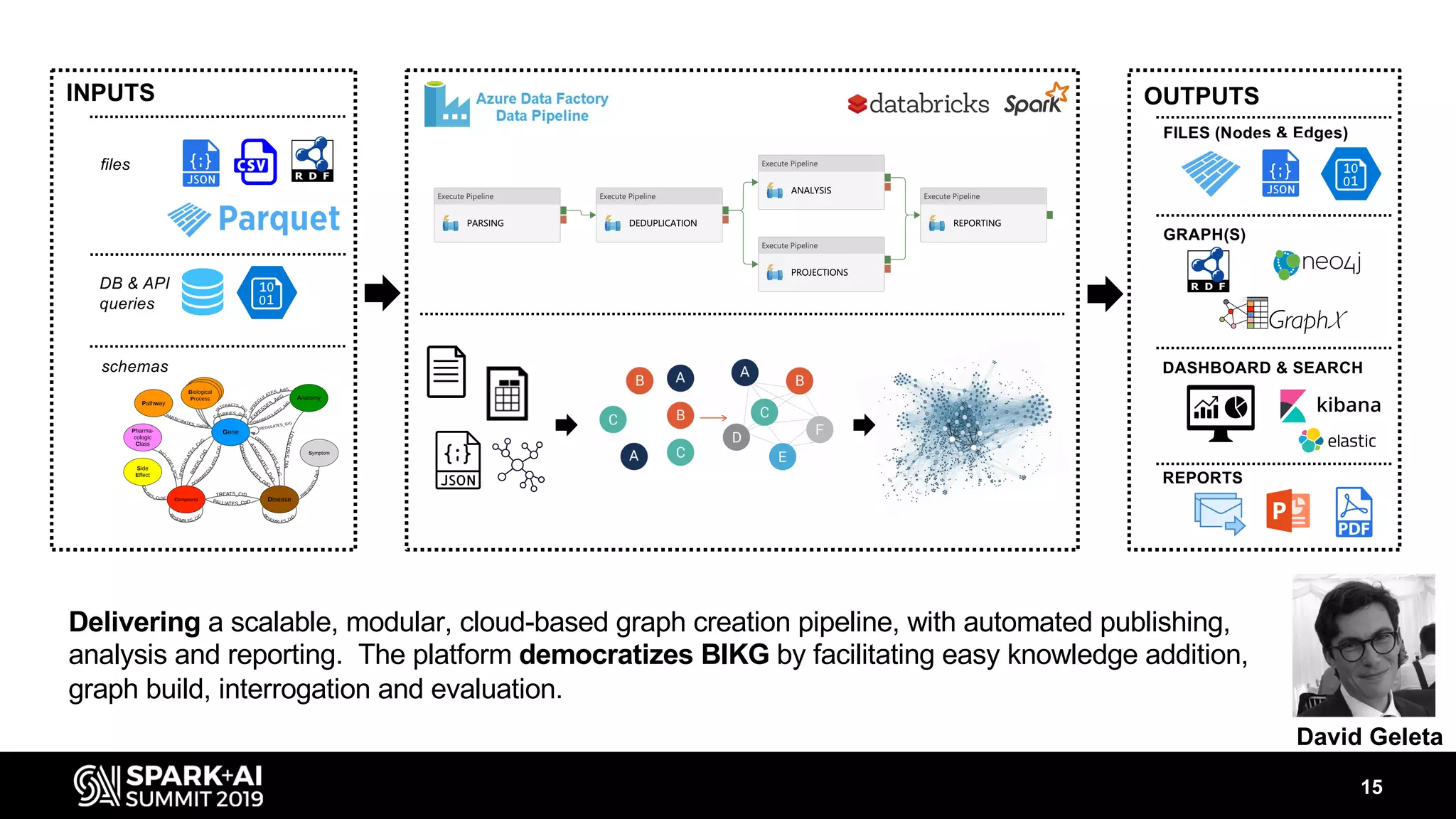
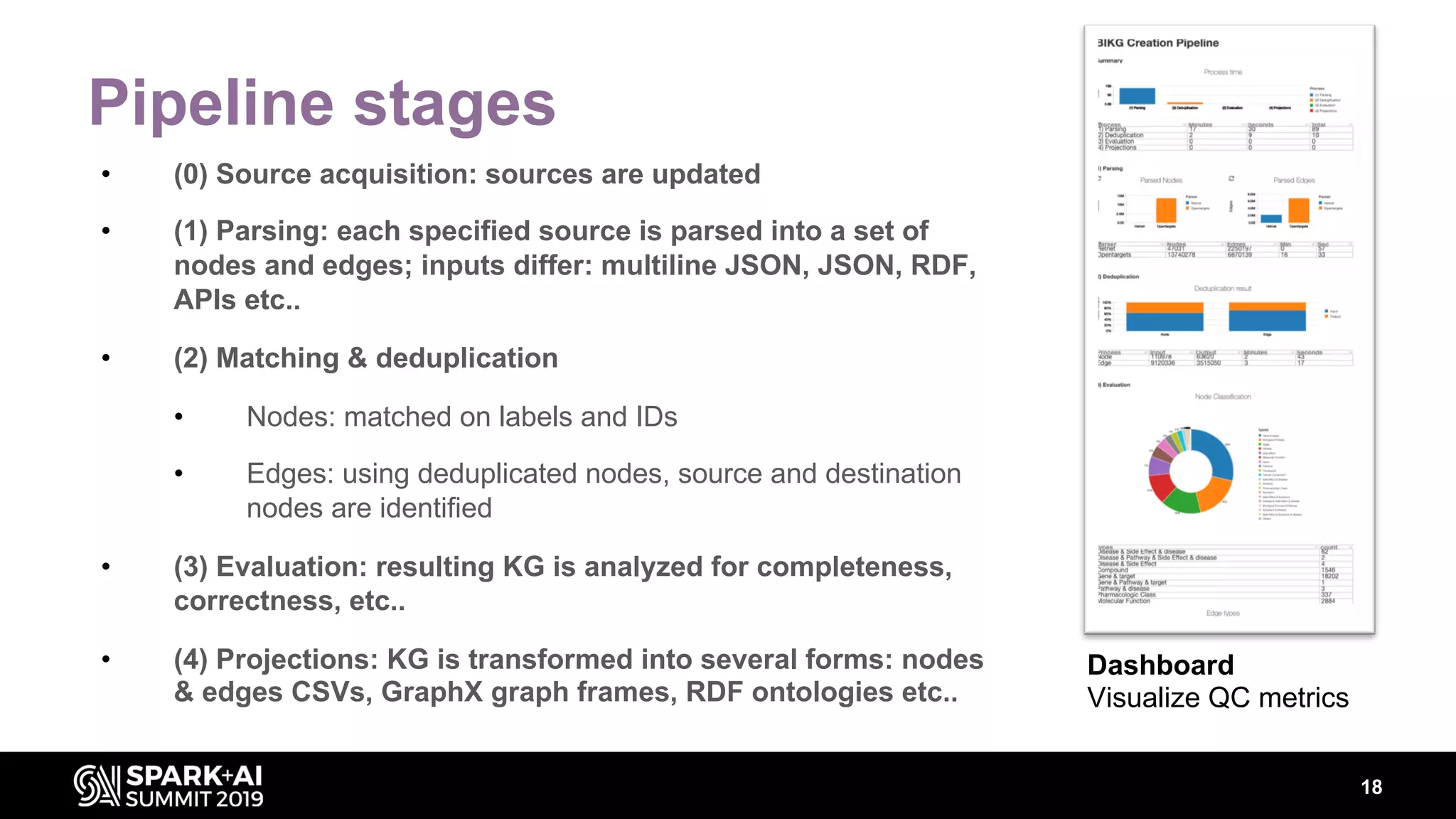
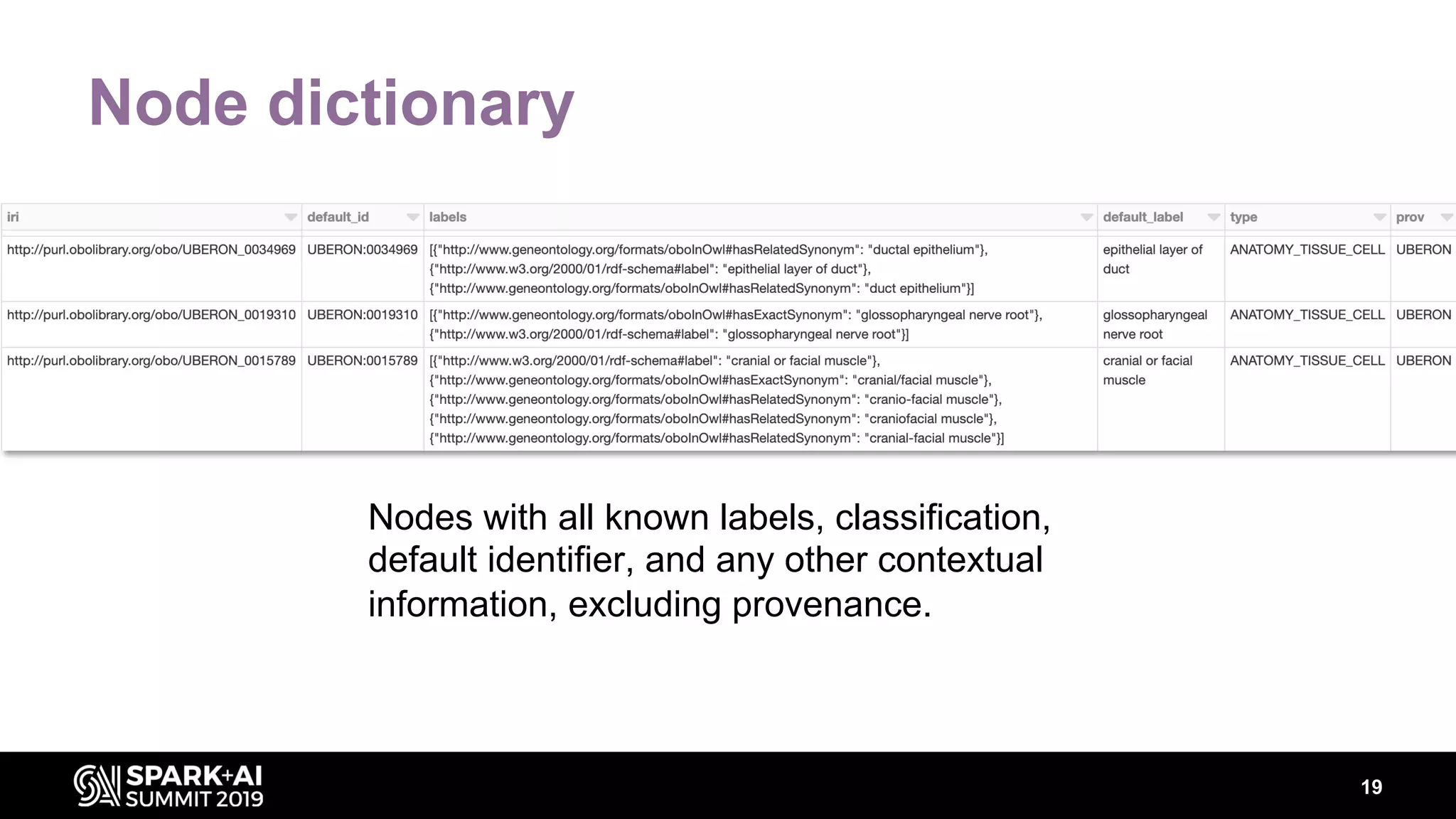
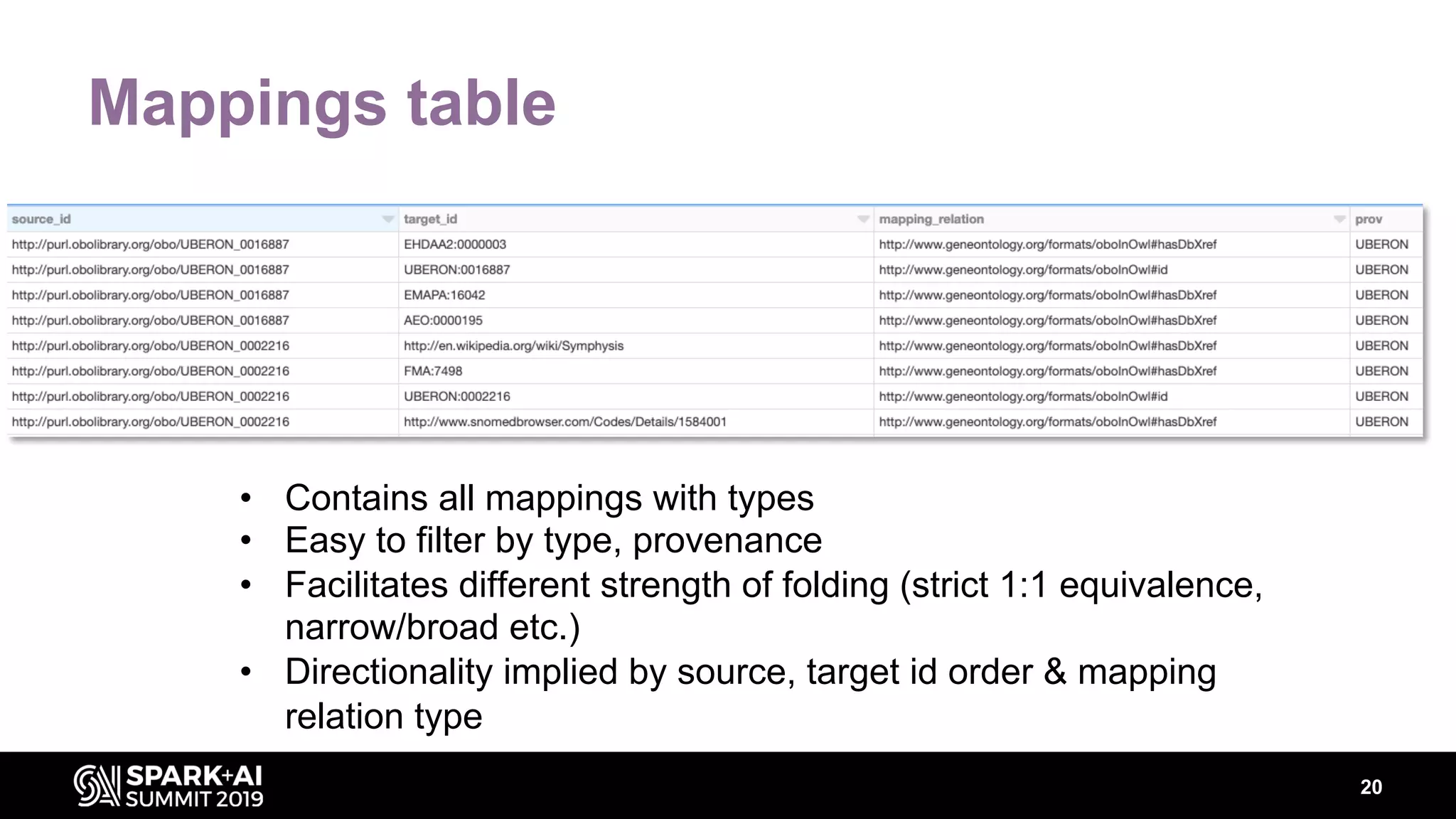
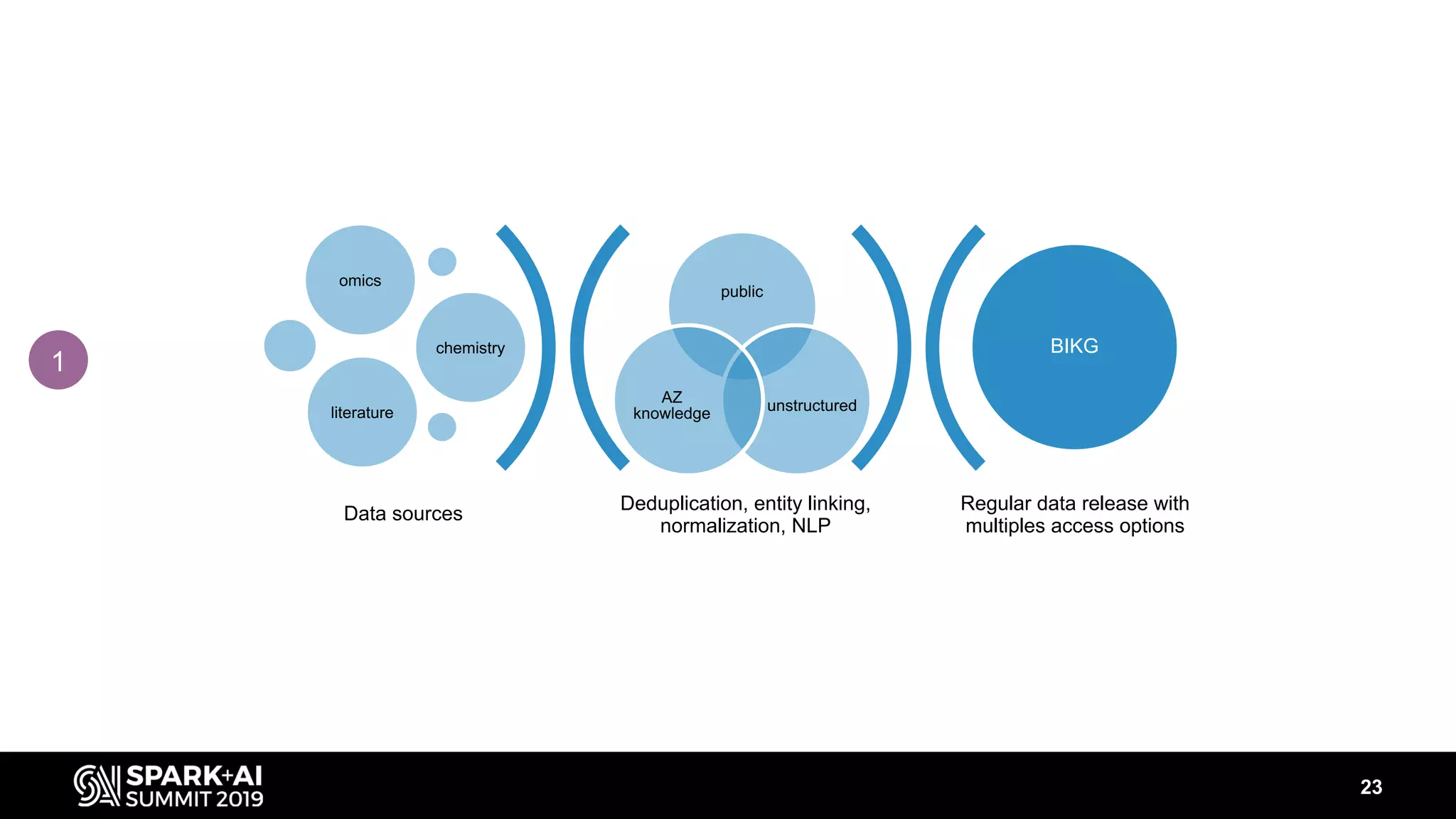
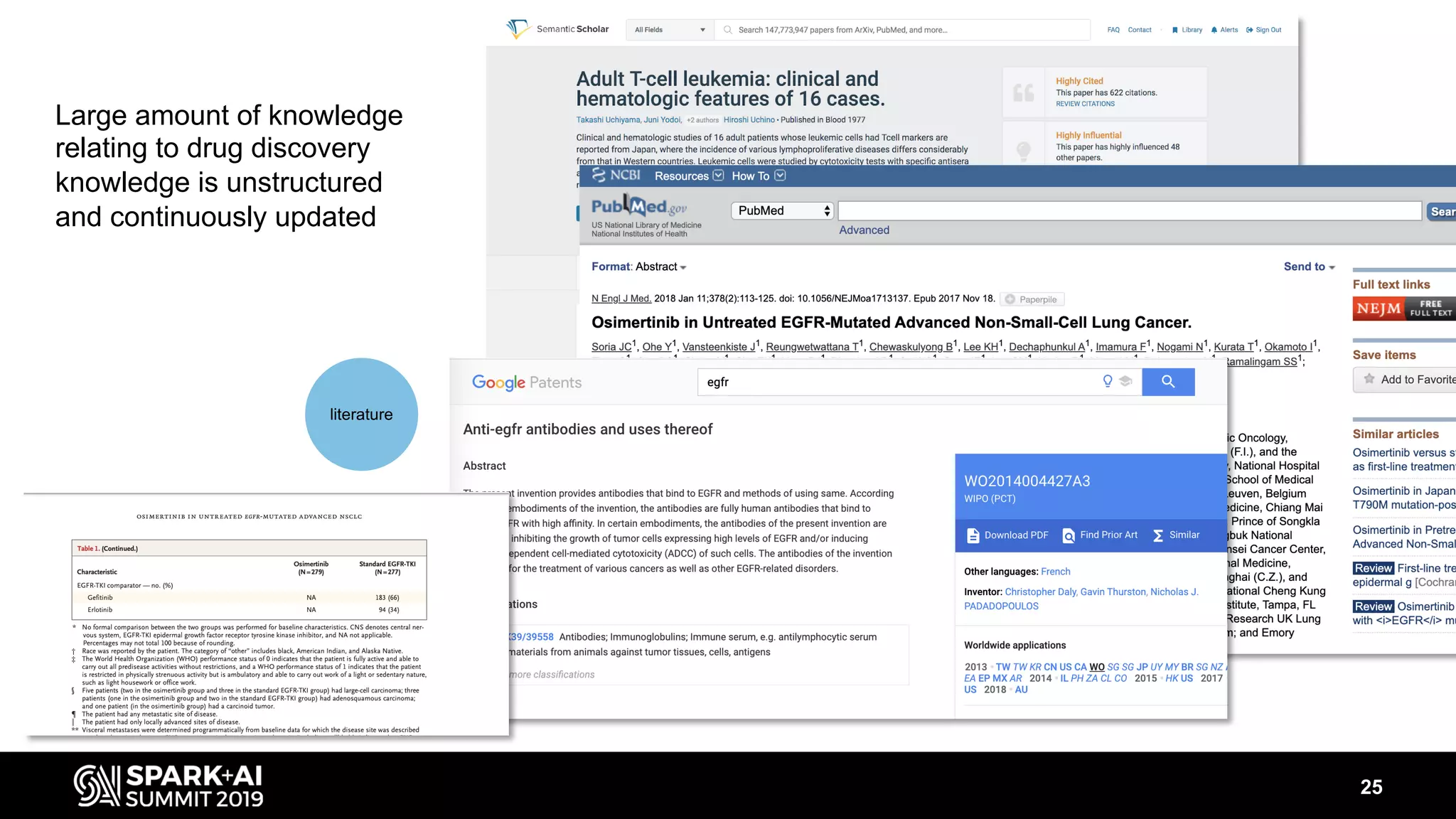
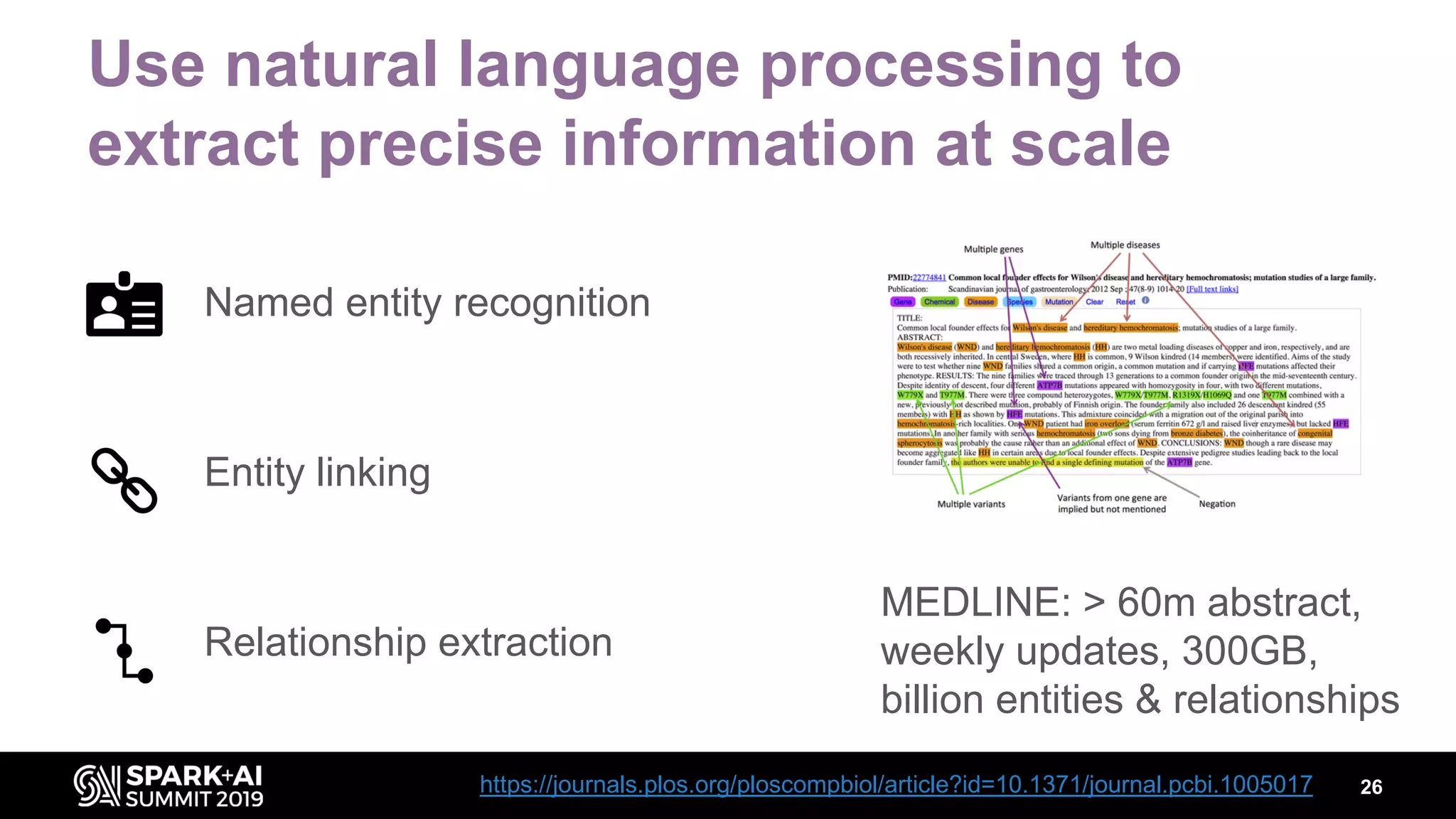
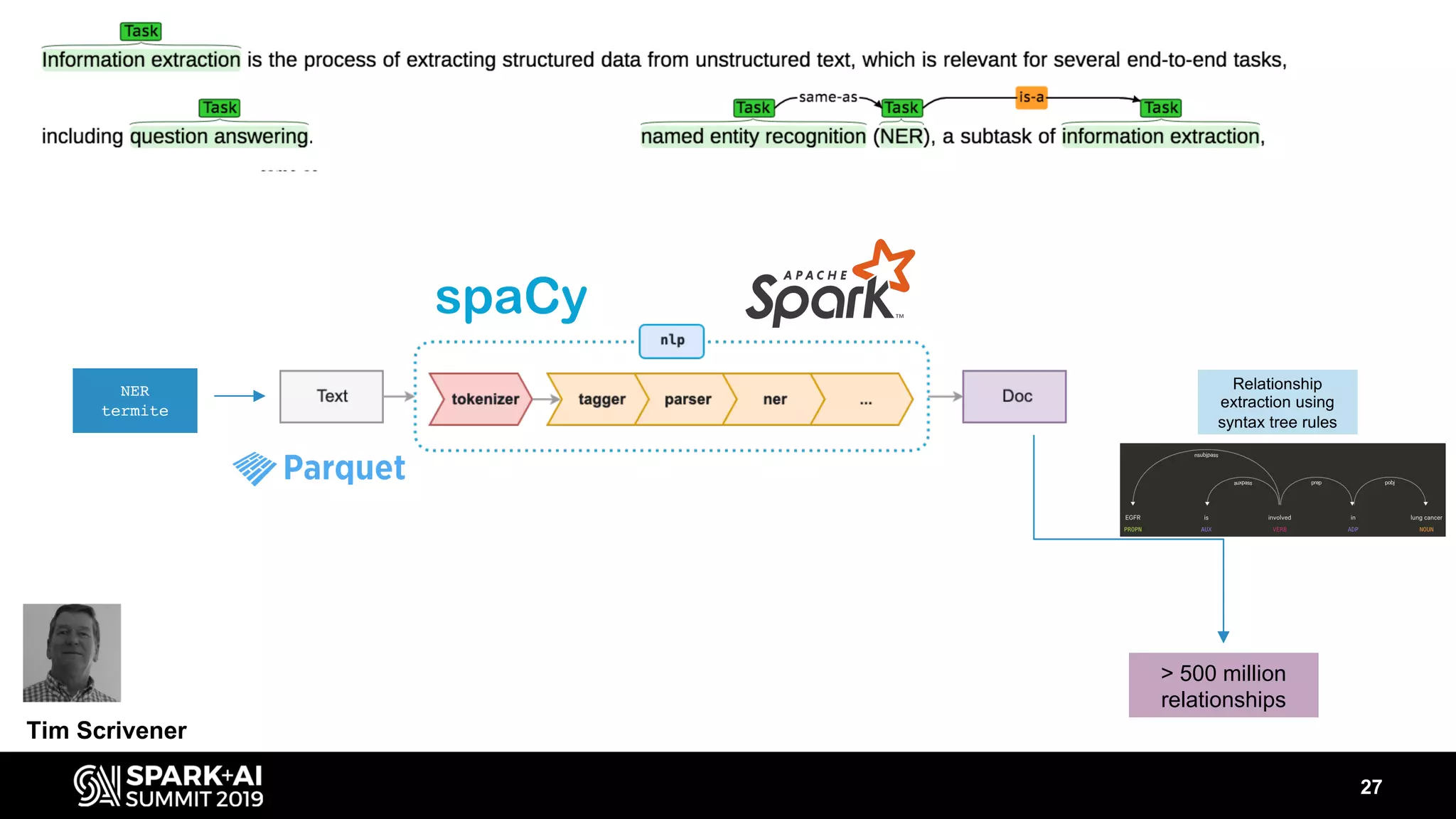
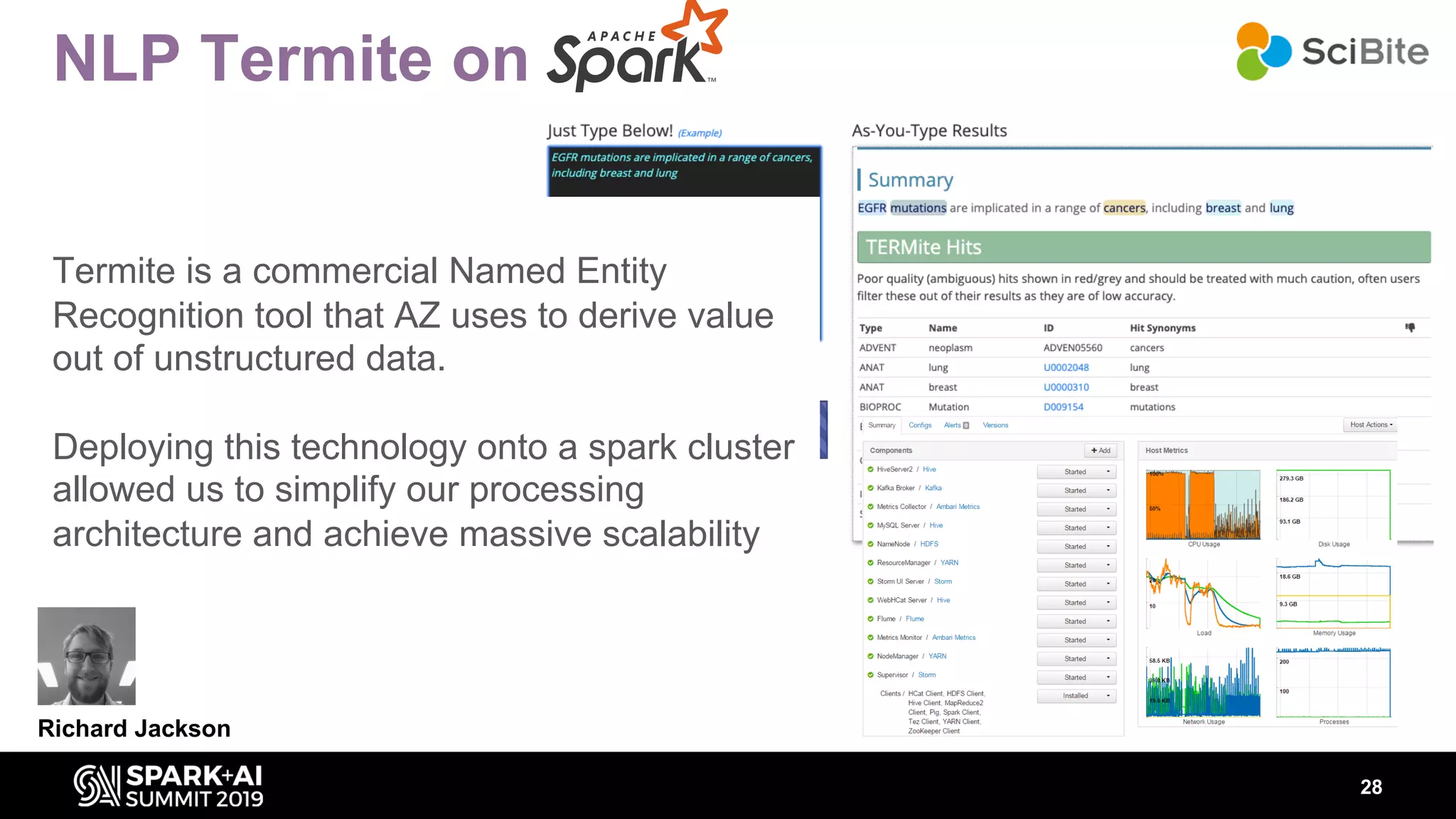
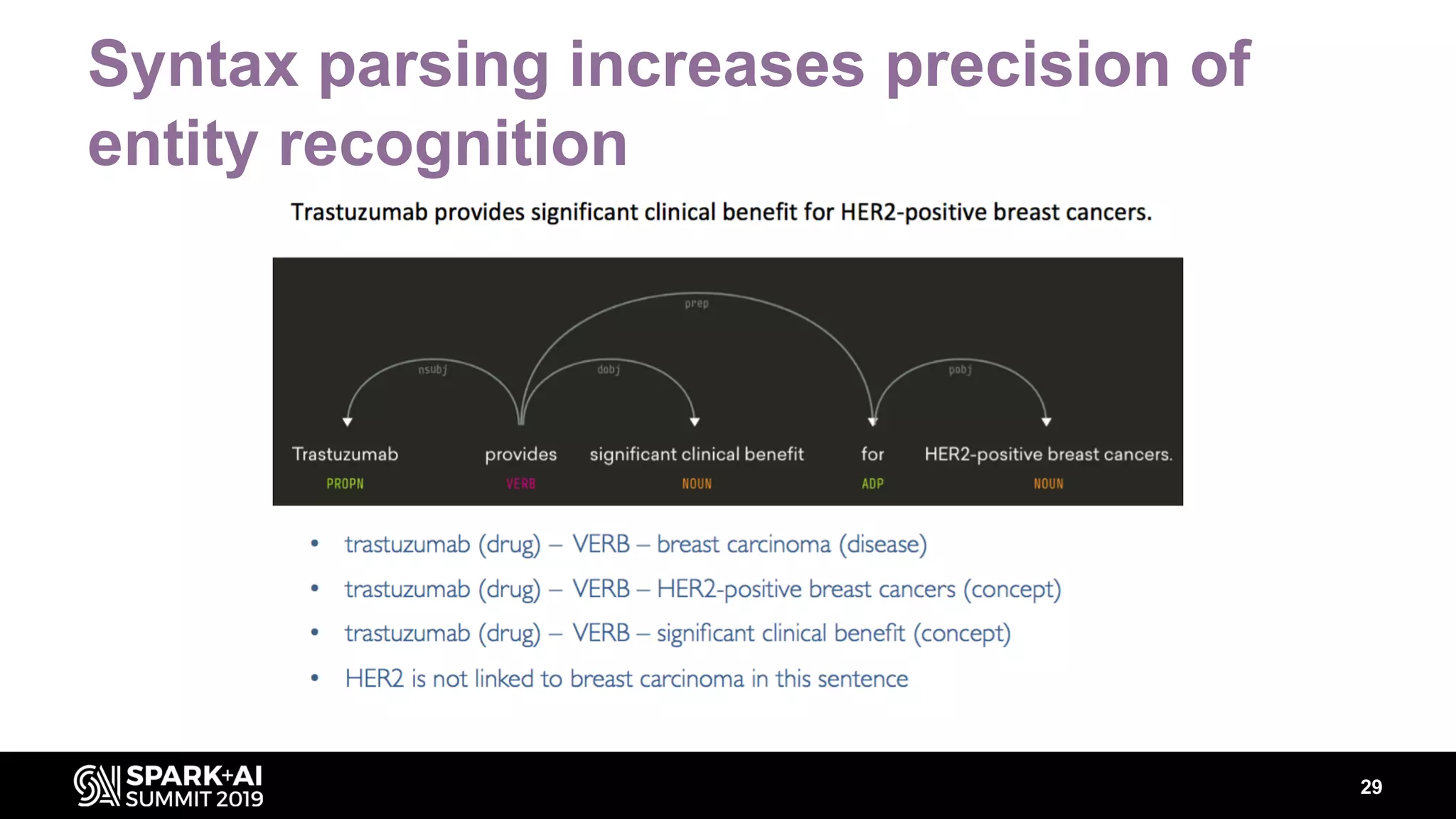
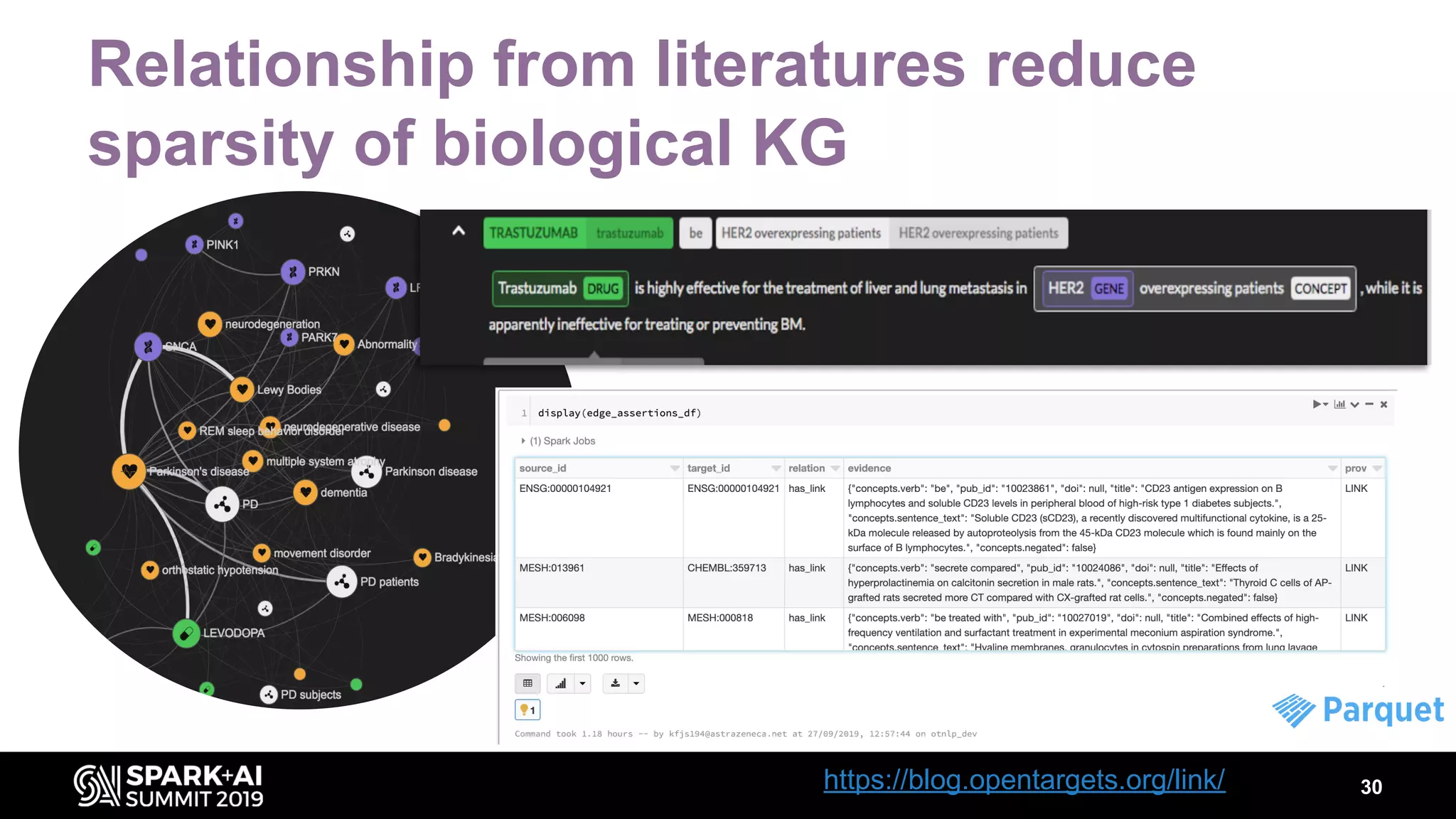
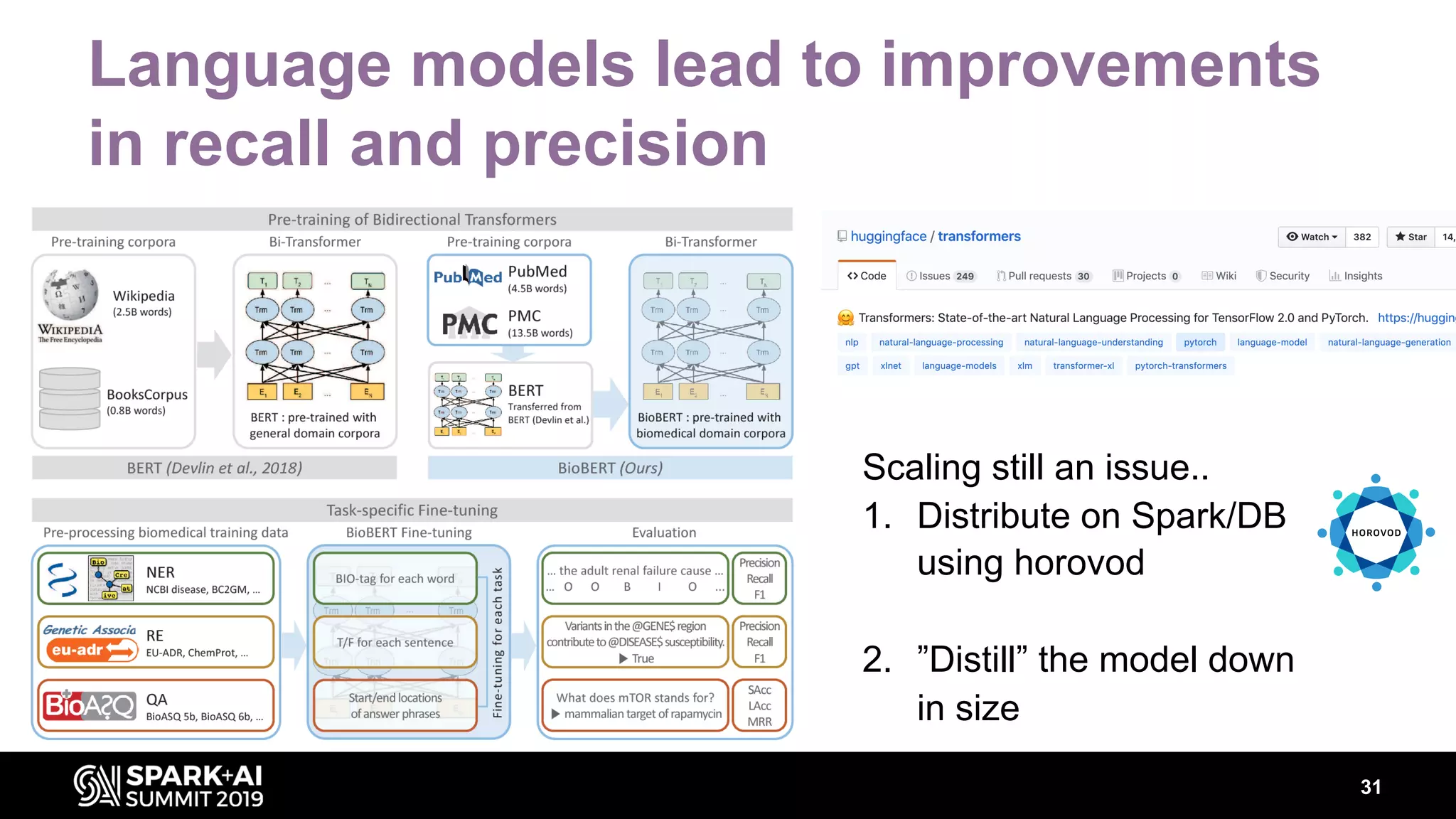
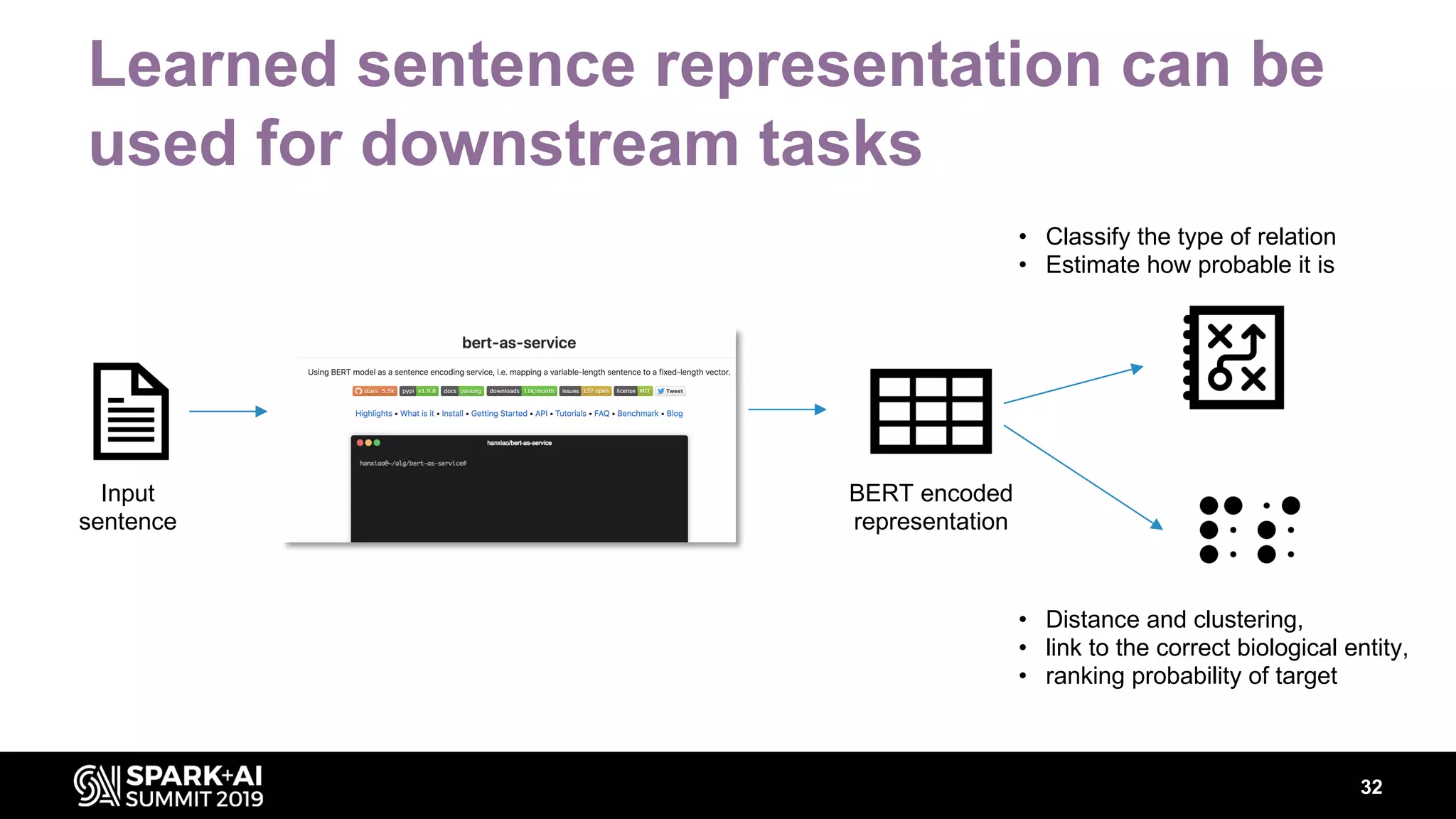
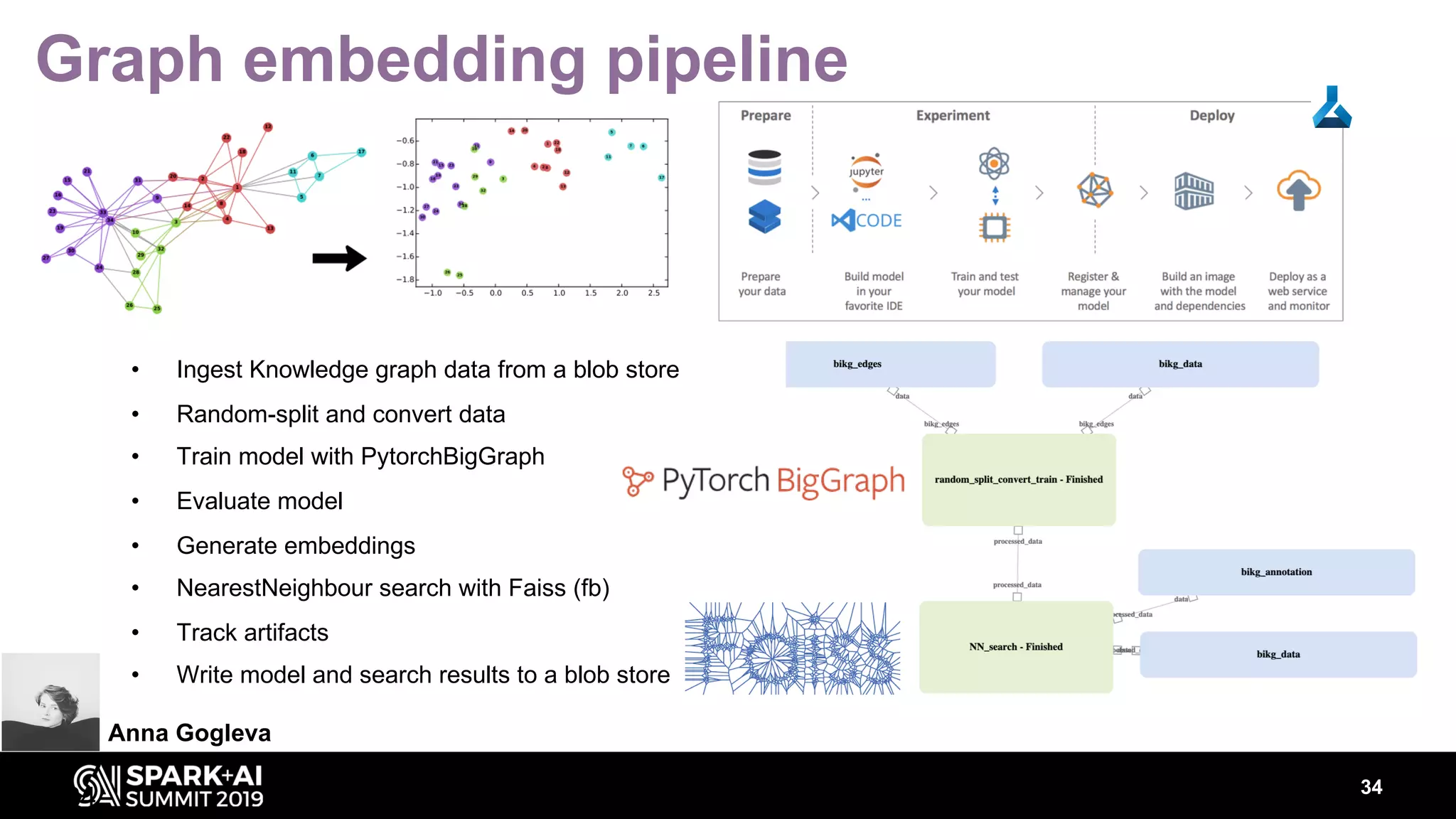
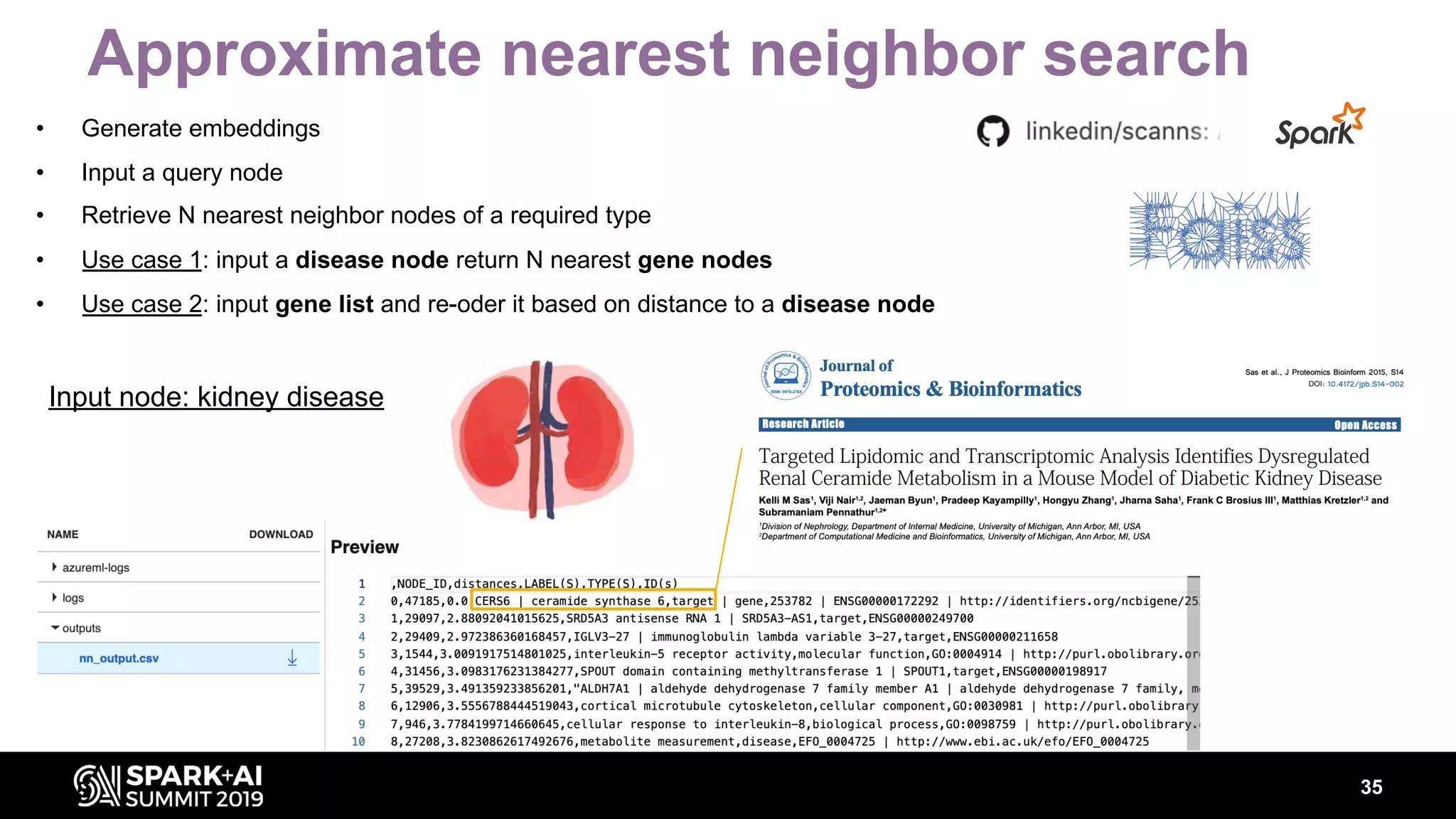
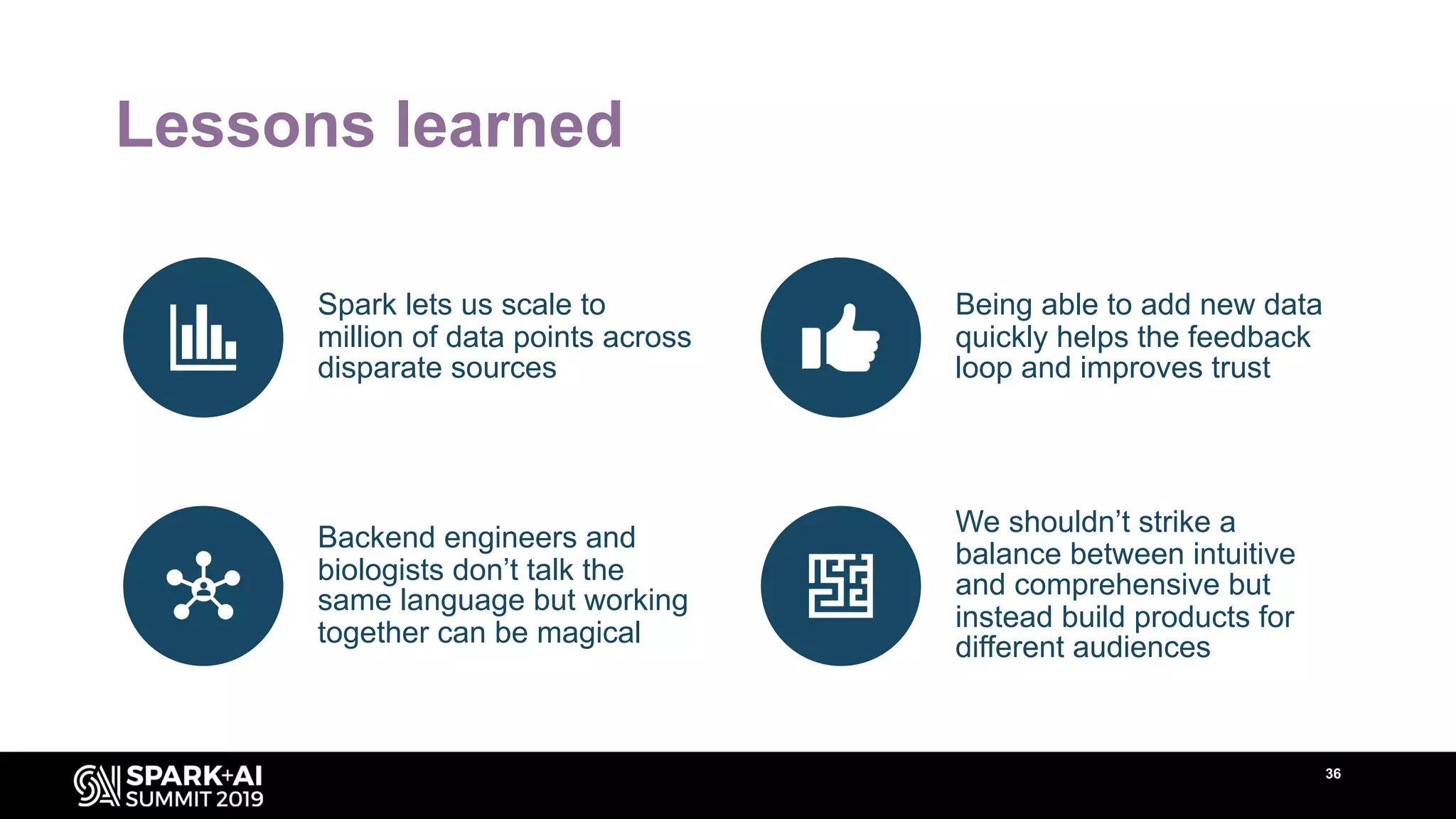
The document discusses AstraZeneca's efforts in utilizing machine learning and natural language processing to enhance drug discovery through the creation of a knowledge graph. It outlines the challenges faced in drug target identification and the proposed '5R' framework to improve decision-making efficiency. Additionally, it details the construction of a scalable cloud-based pipeline for knowledge extraction and the integration of various data sources to aid scientists in generating hypotheses.