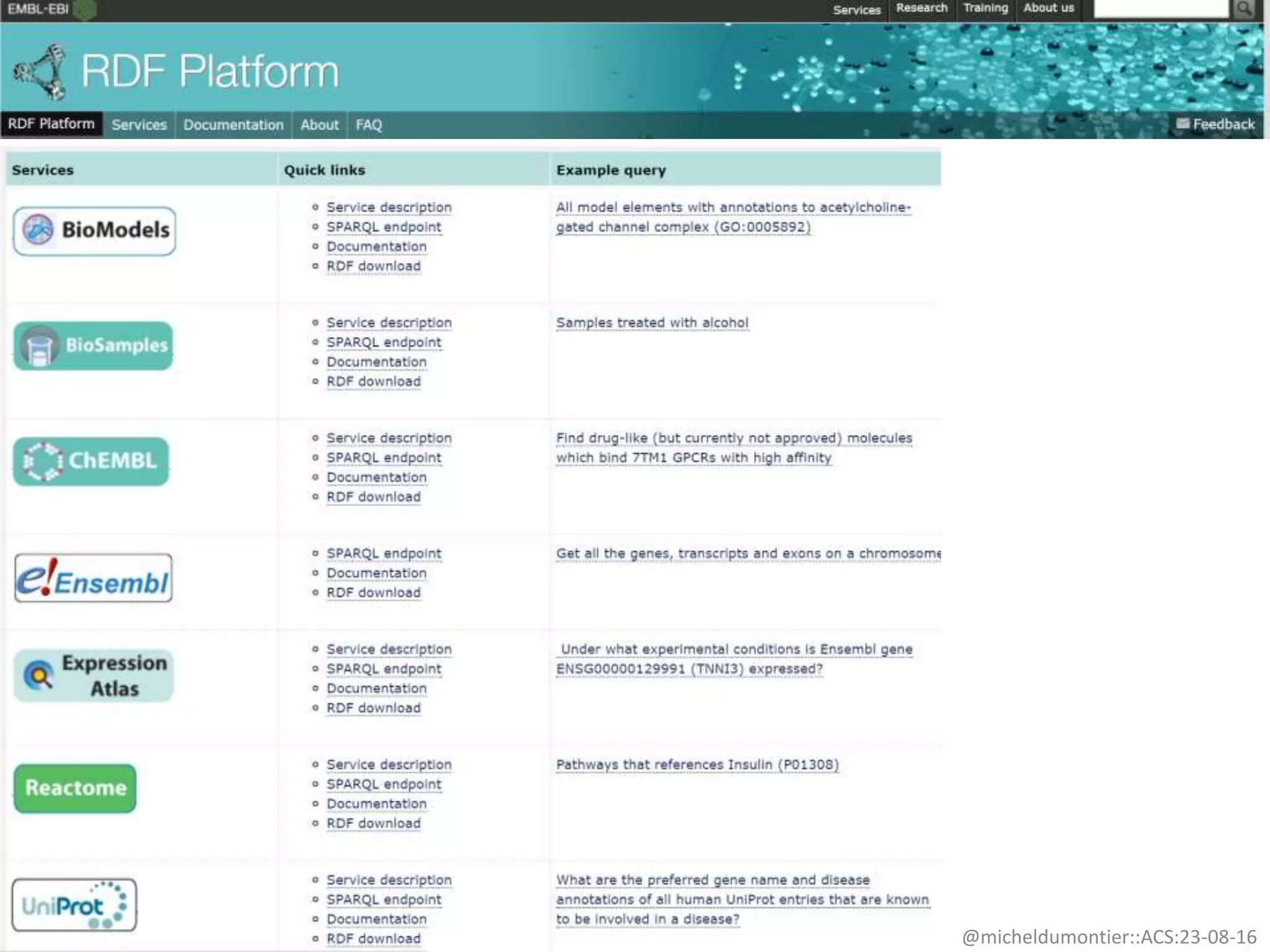
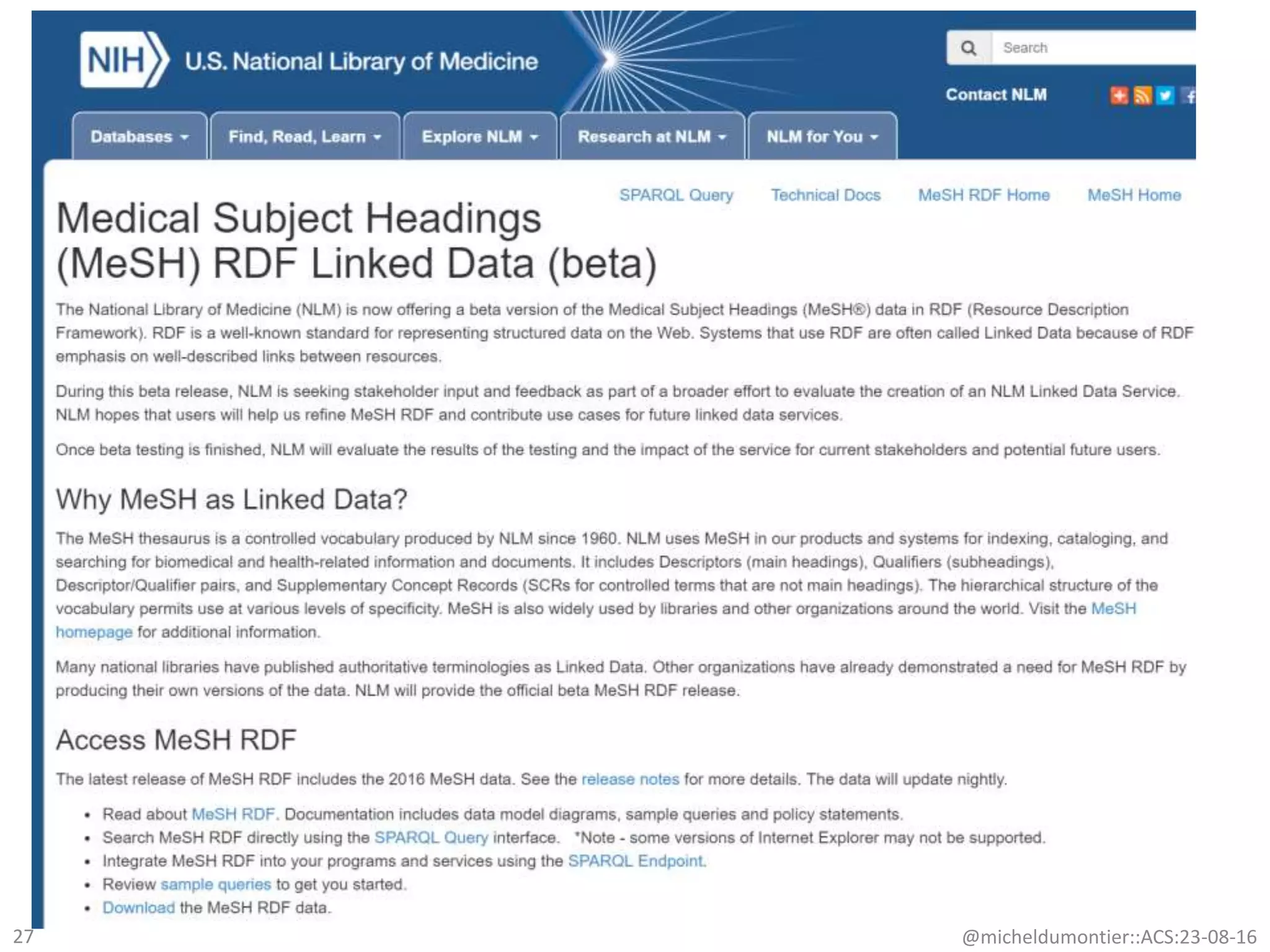
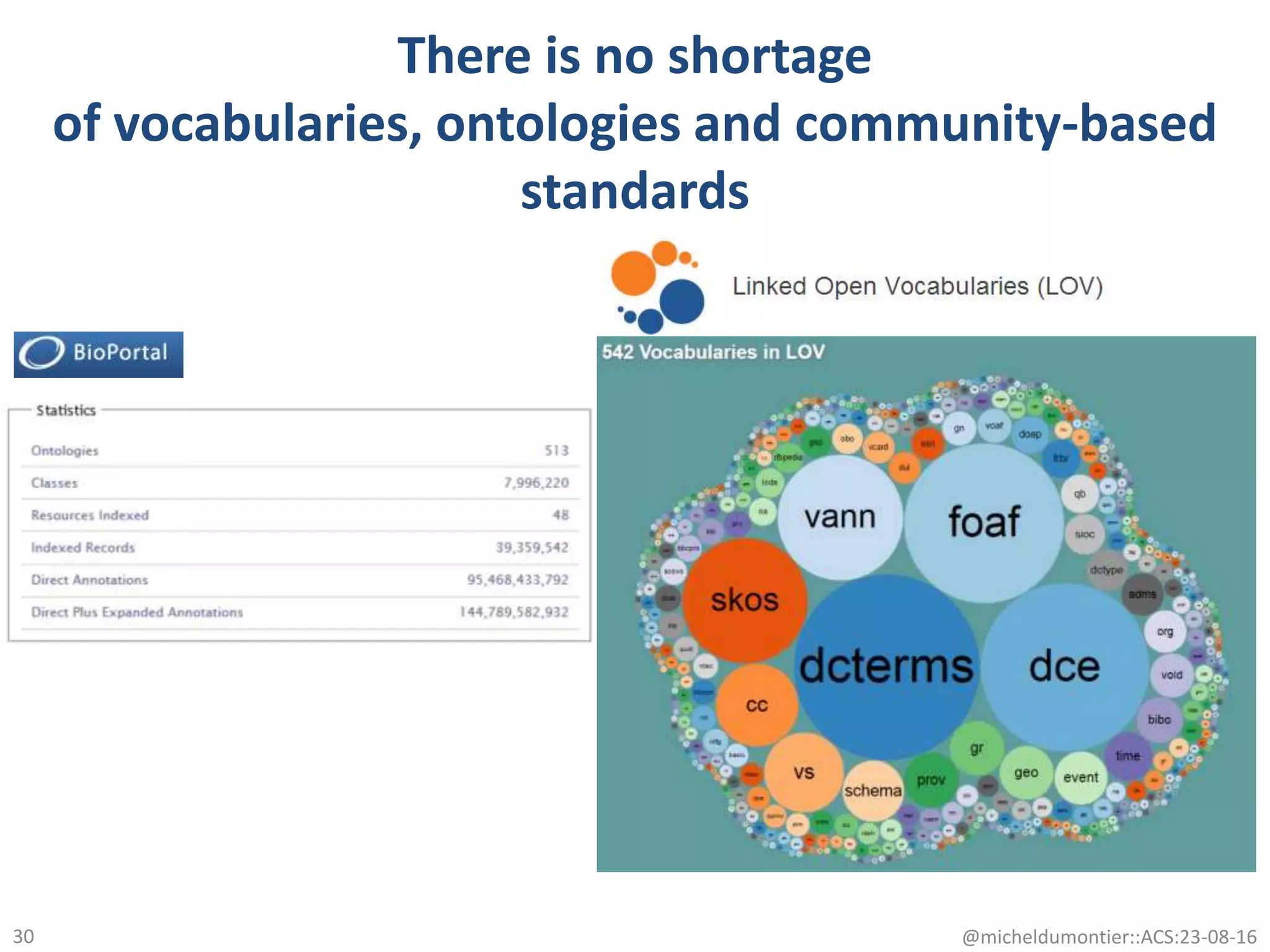
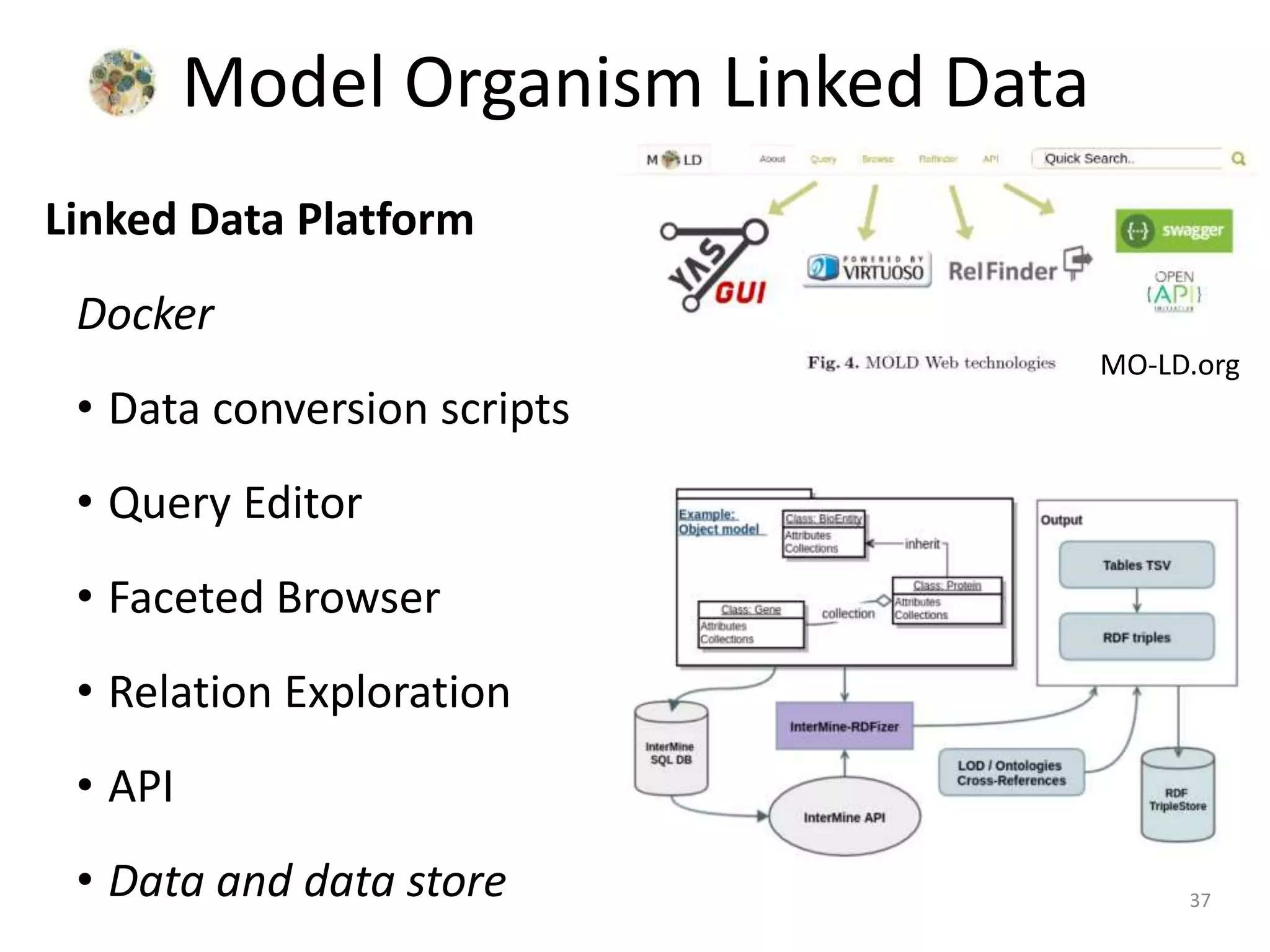
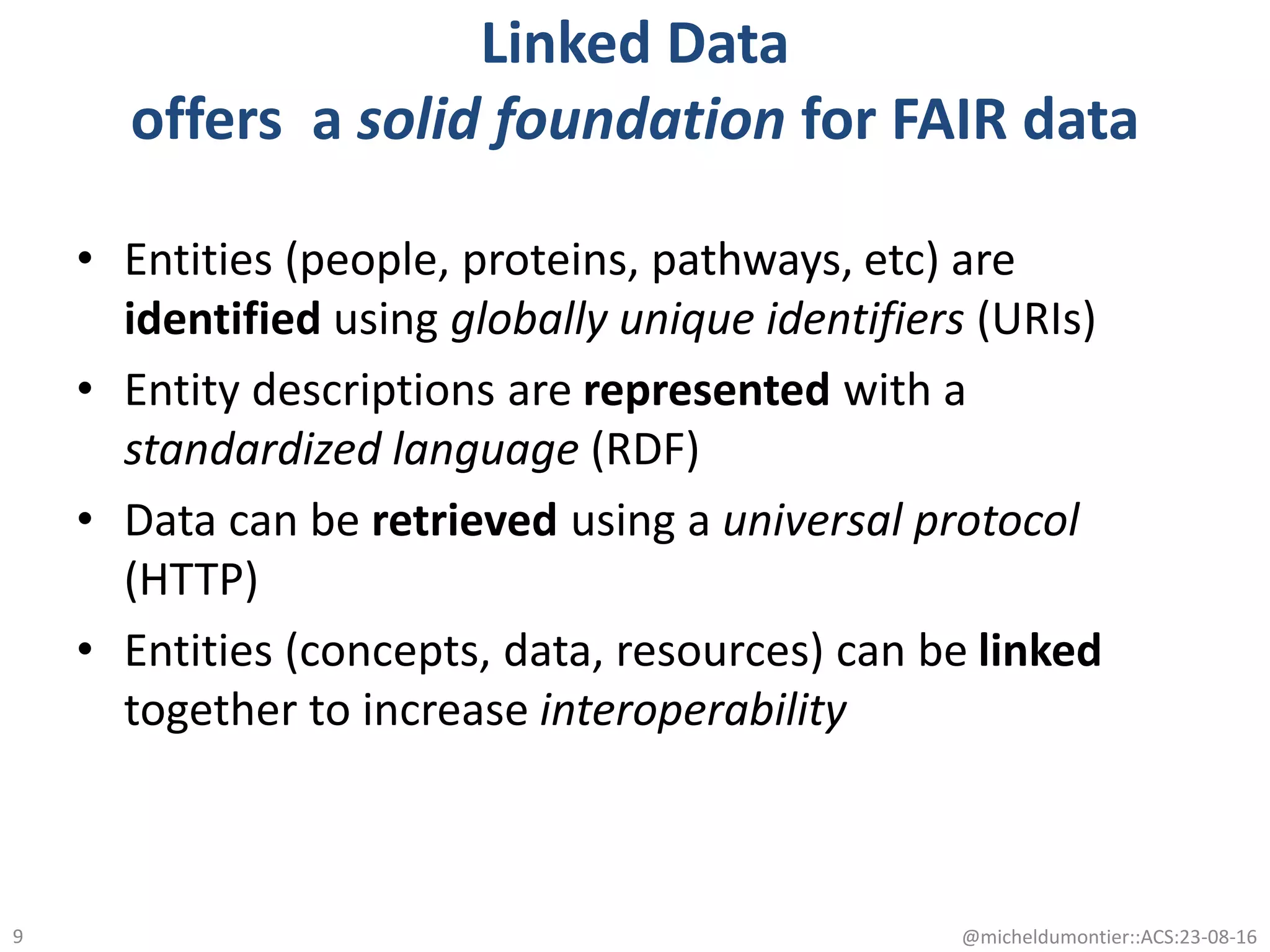
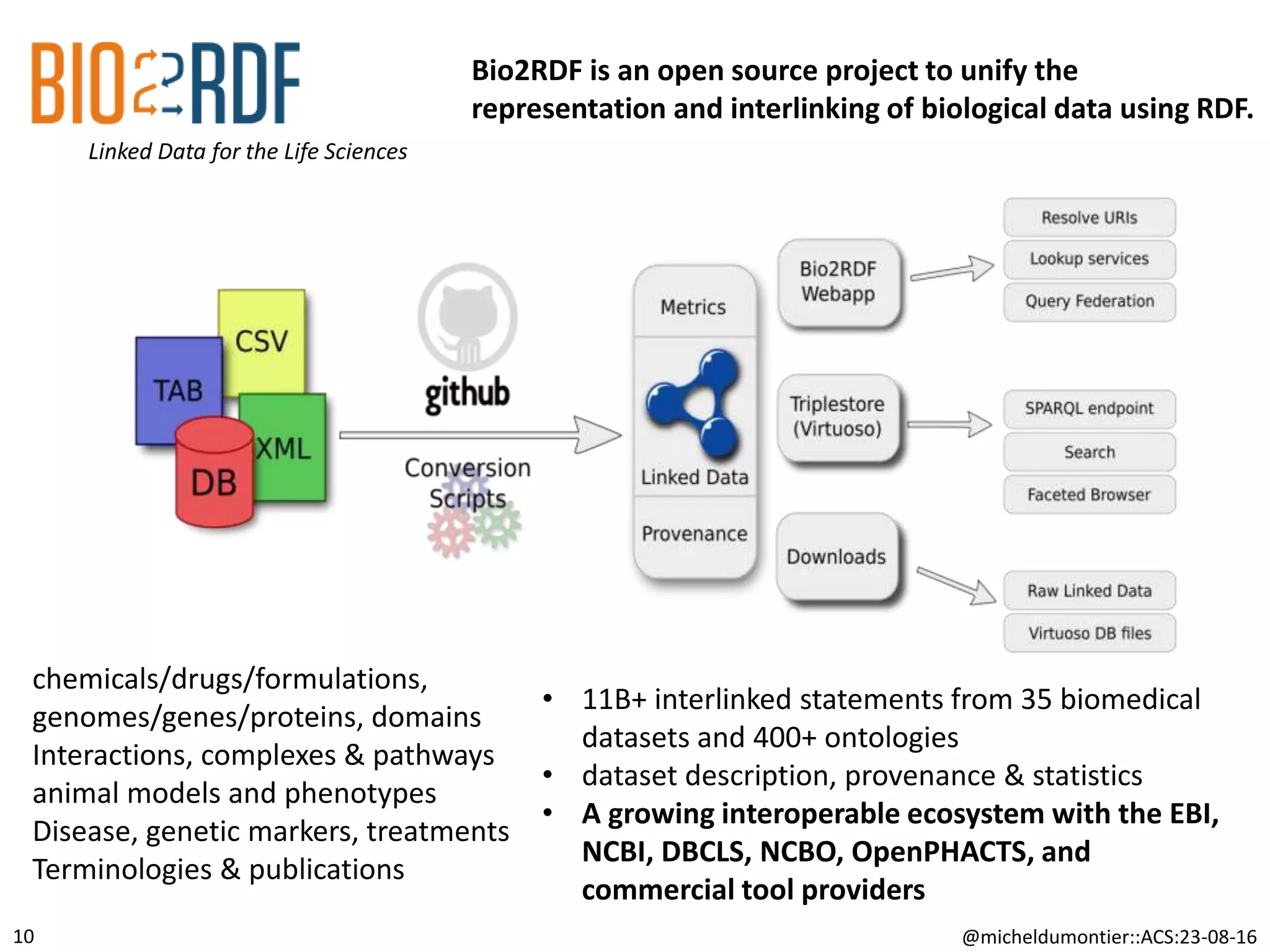
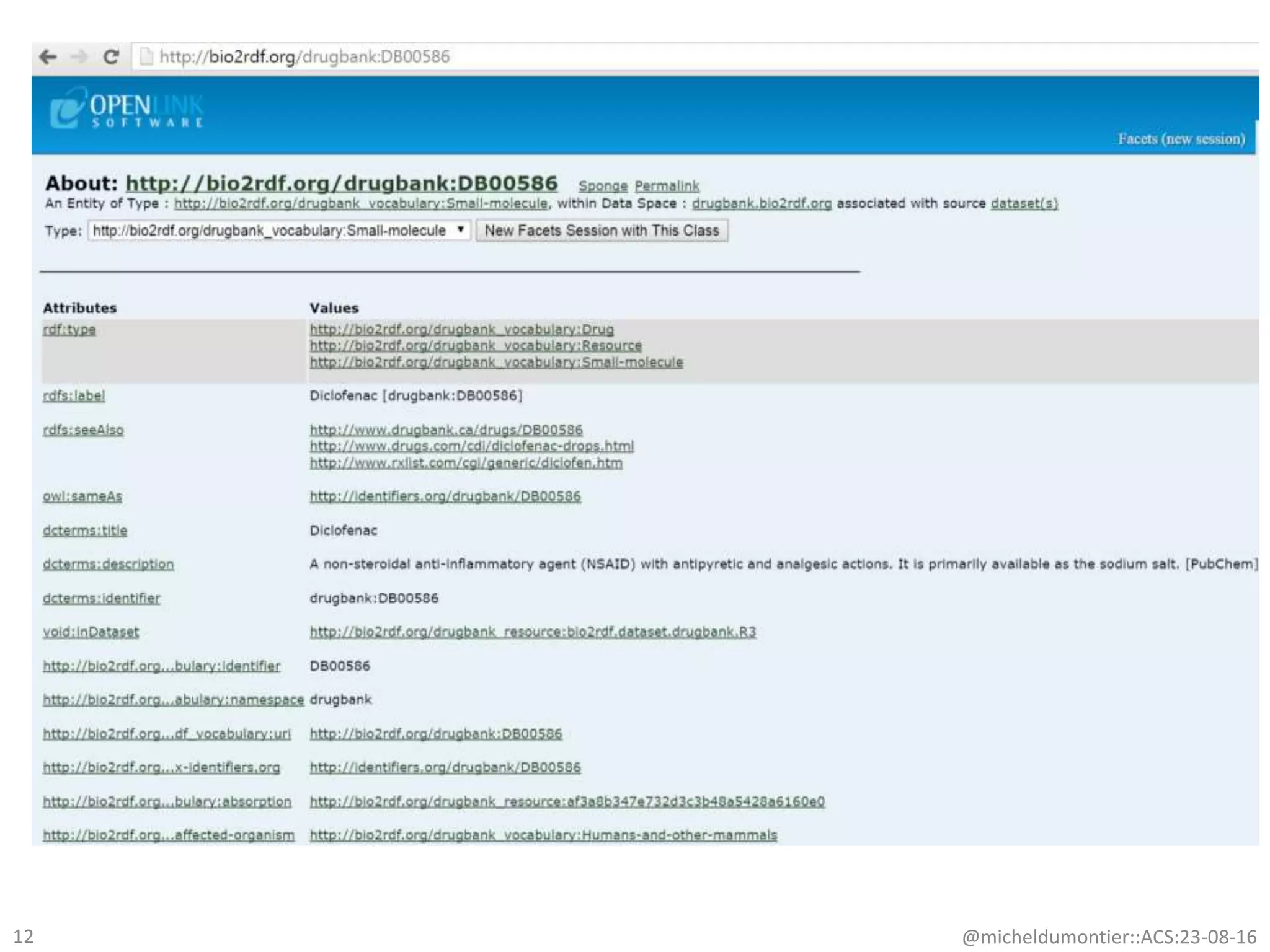
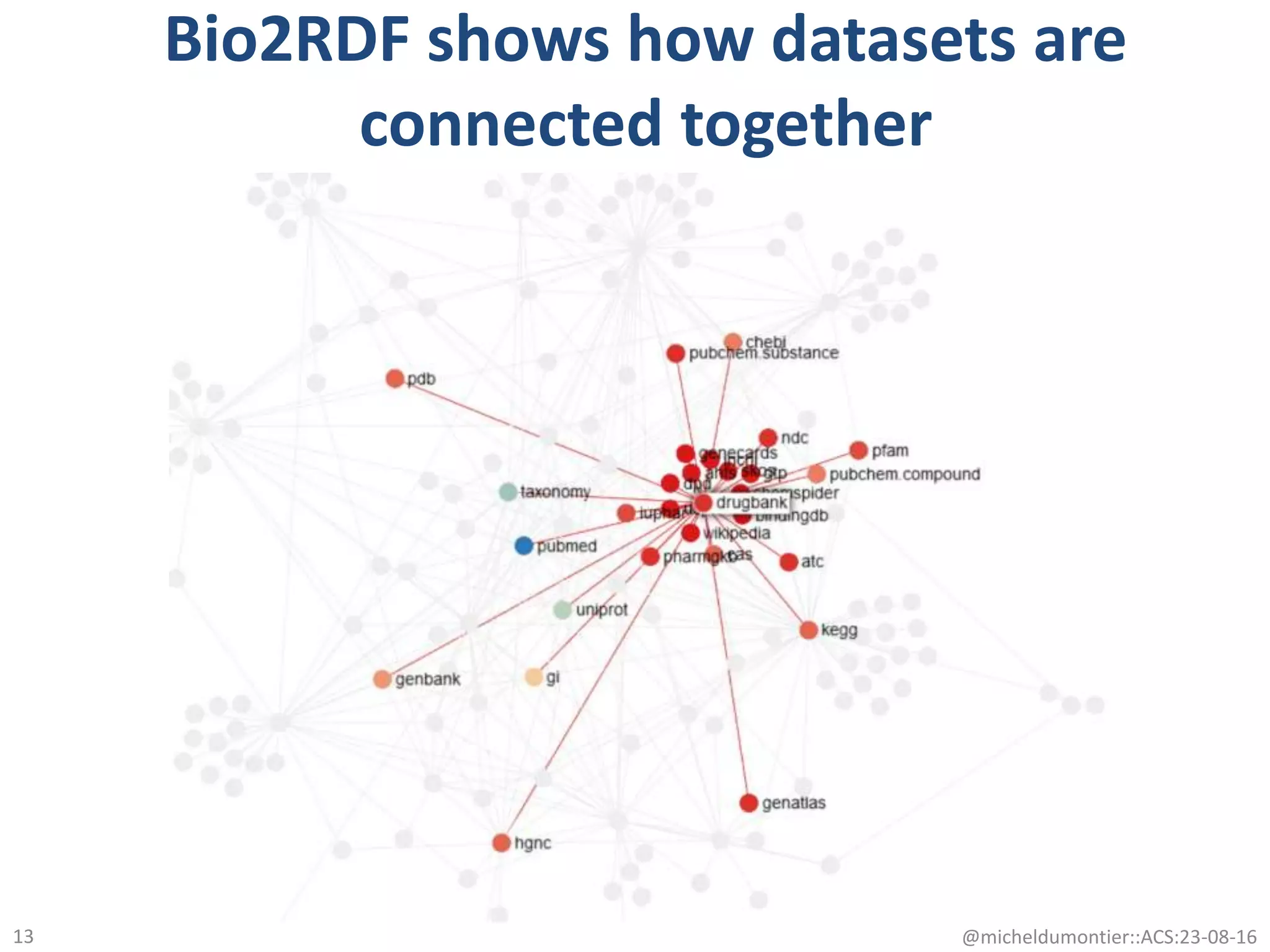
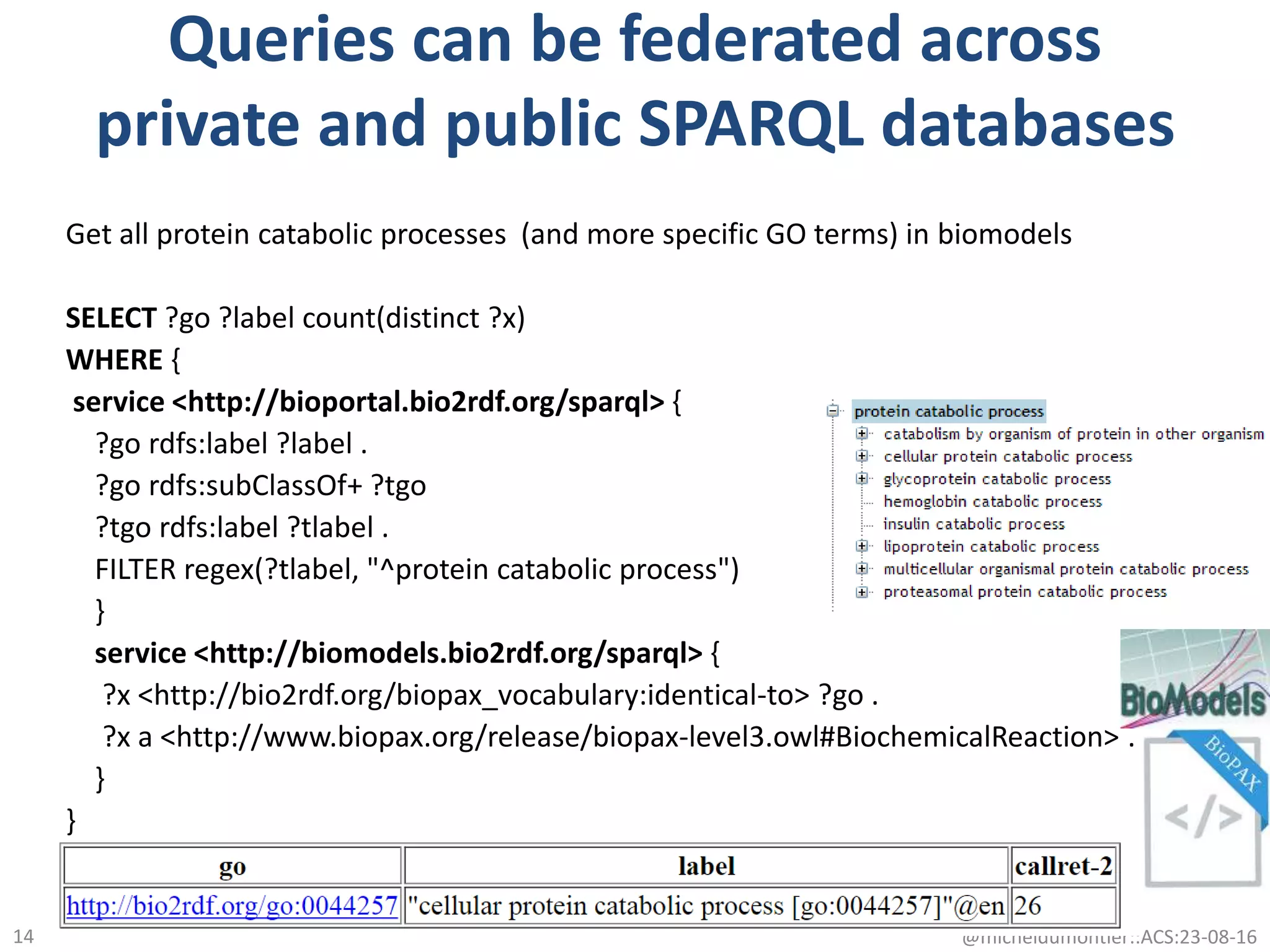
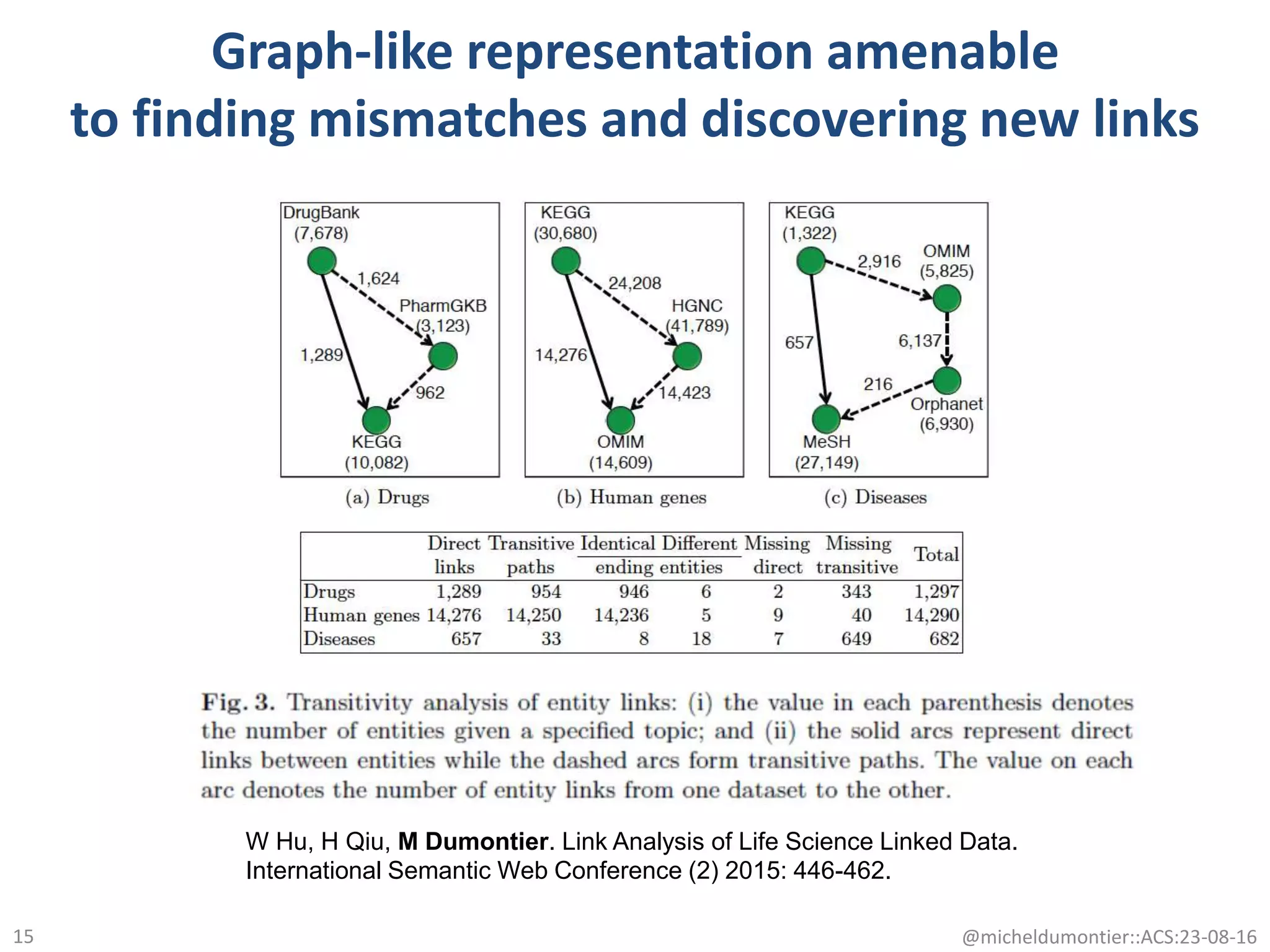
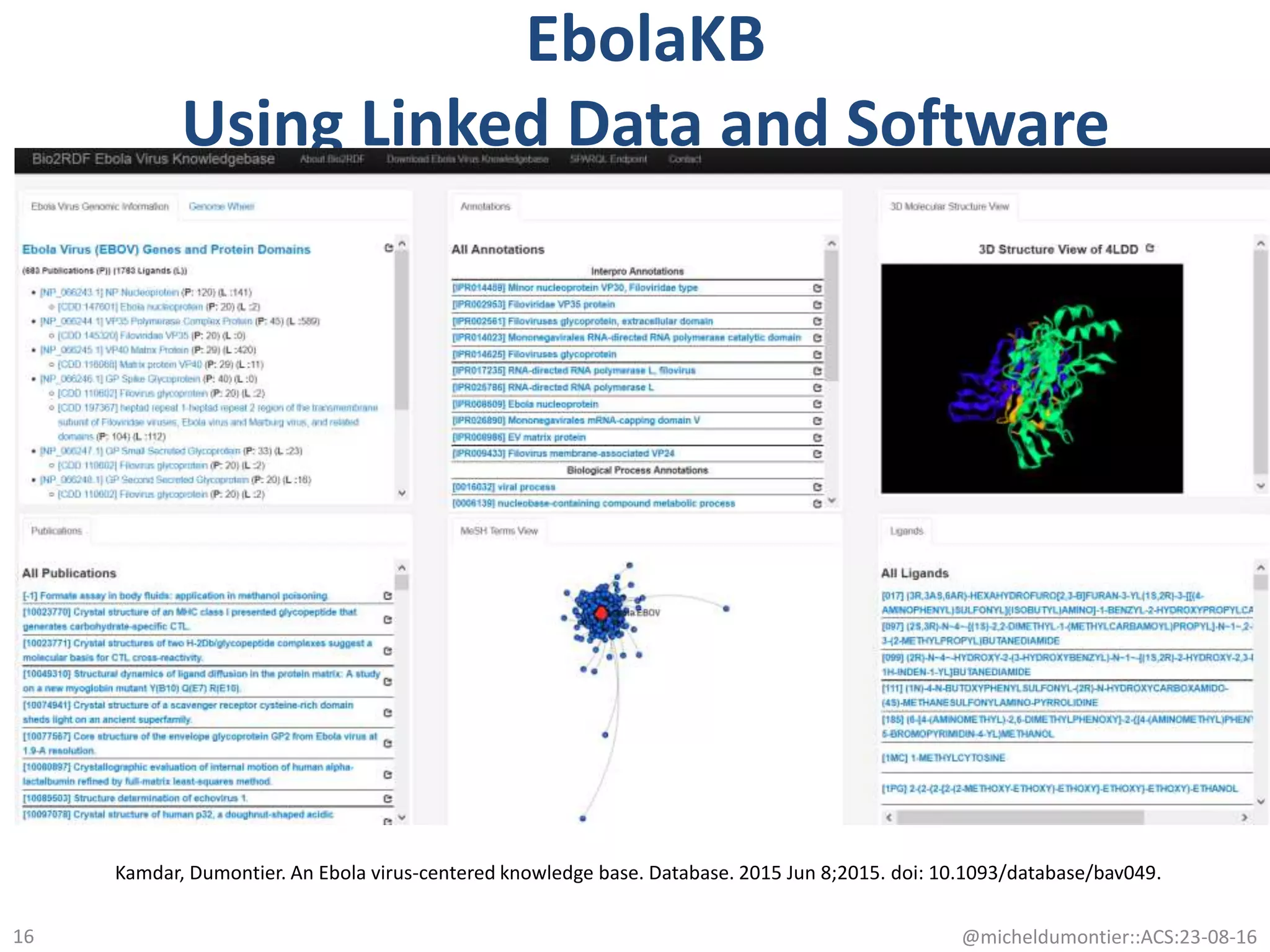
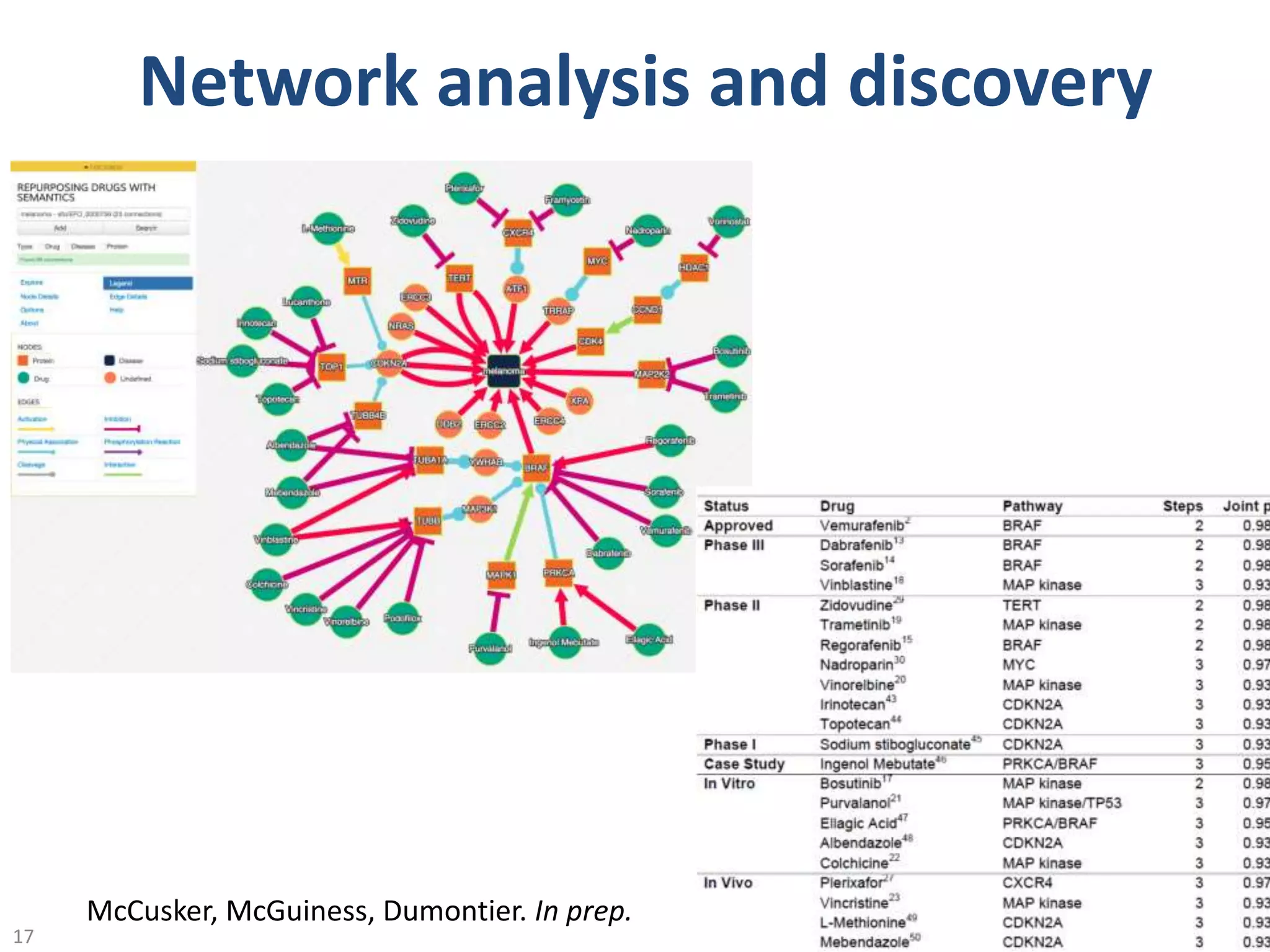
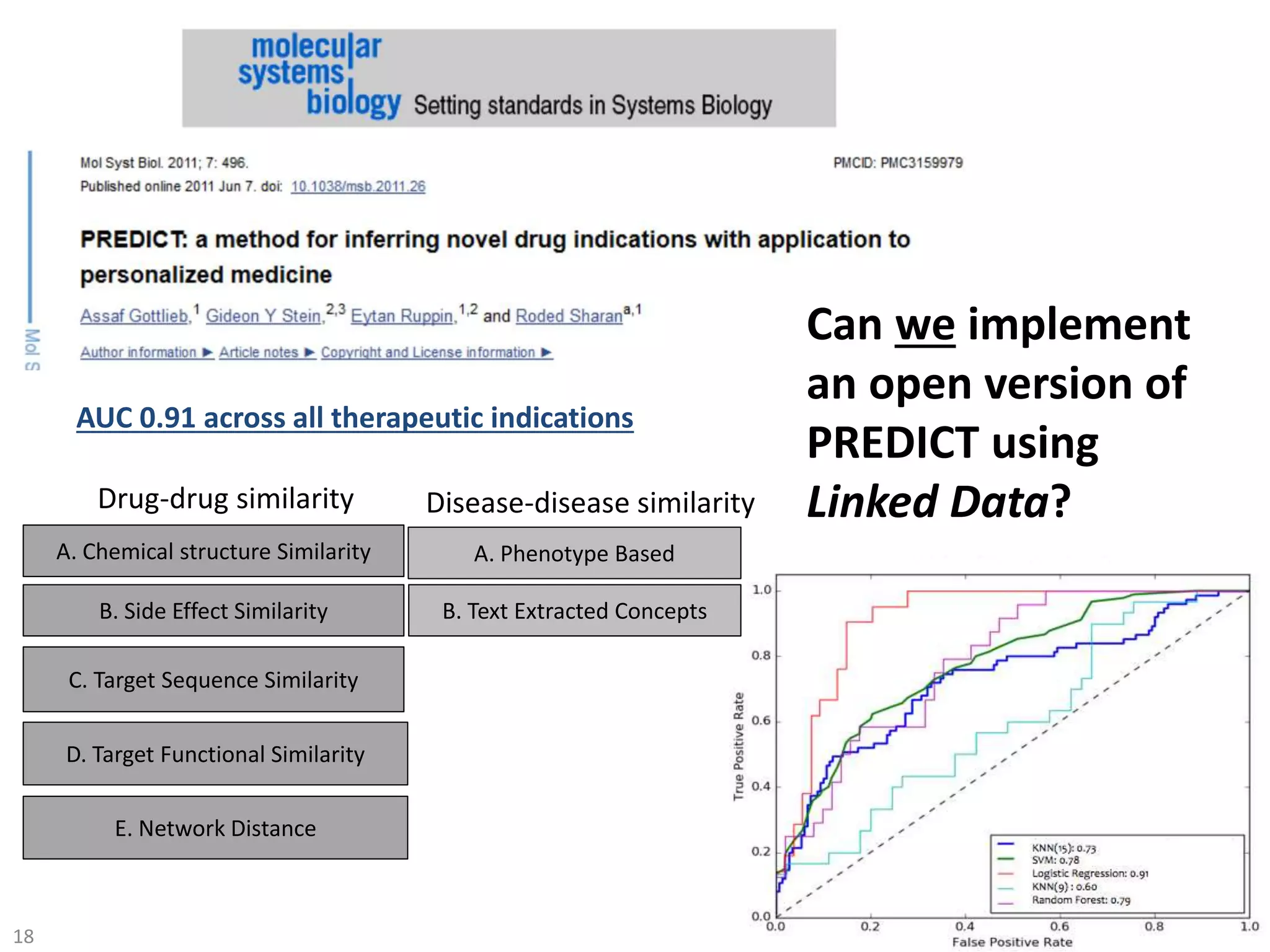
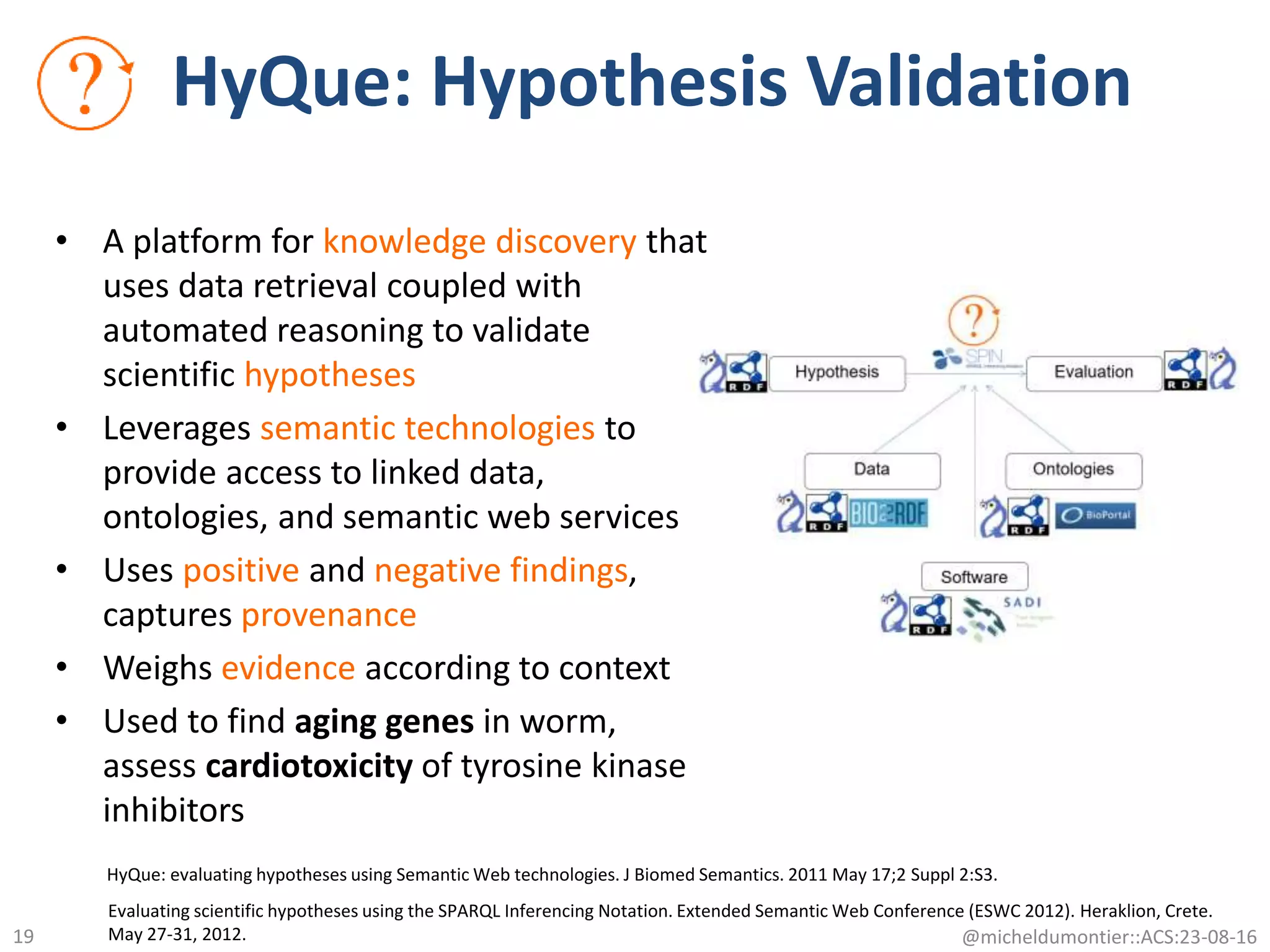
The document discusses the development of a network of interoperable linked and open biomedical data to enhance knowledge discovery and reproducibility in research. It emphasizes the importance of standards and methods for publishing, sharing, and querying biomedical data while highlighting projects like Bio2RDF that unify biological data representation. The content stresses the need for ongoing efforts to ensure data reusability and improve access to interconnected biomedical resources.






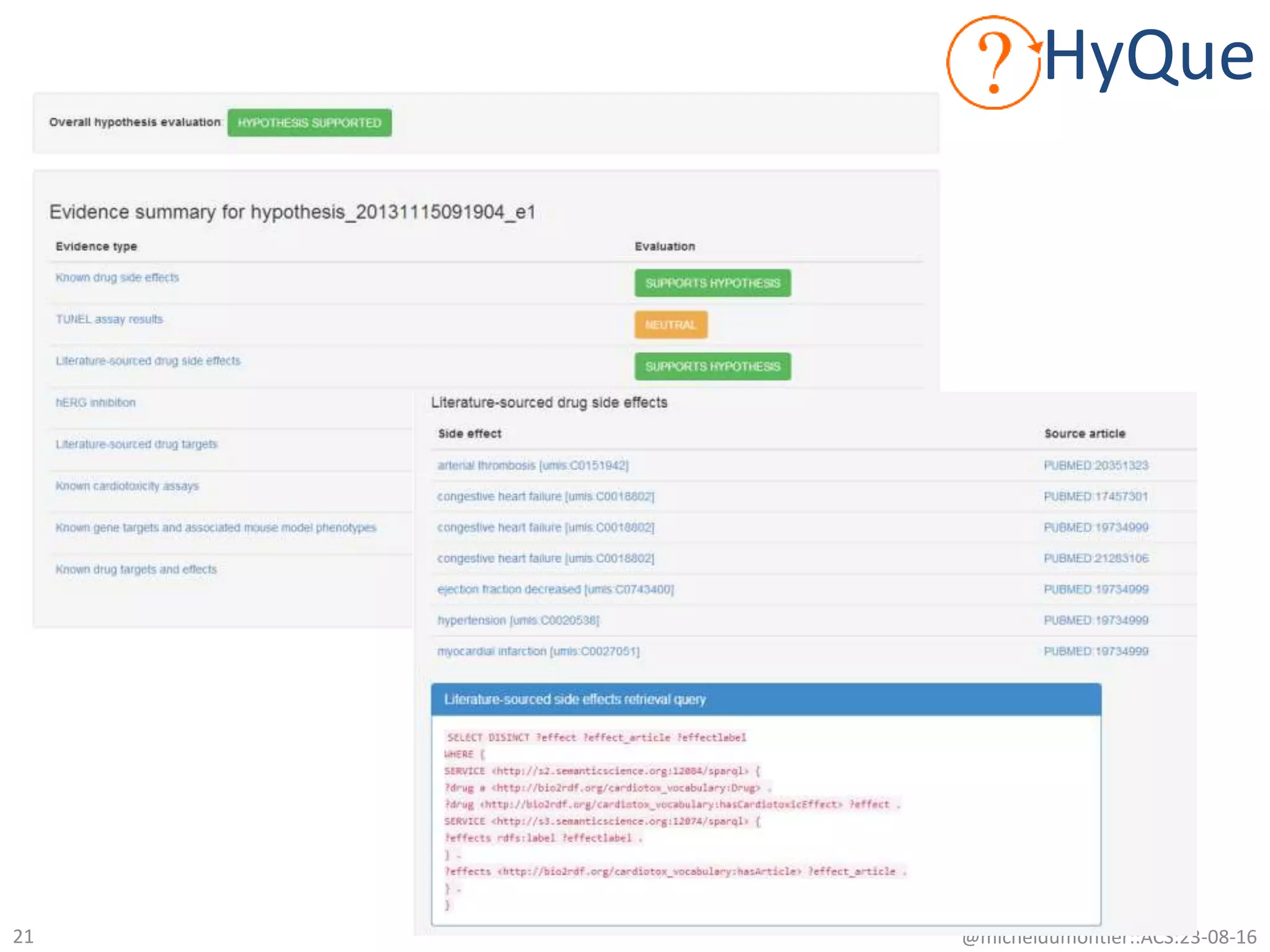
![What evidence might we gather?
• clinical: Are there cardiotoxic effects associated with the drug?
– Literature (studies) [curated db]
– Product labels (studies) [r3:sider]
– Clinical trials (studies) [r3:clinicaltrials]
– Adverse event reports [r2:pharmgkb/onesides]
– Electronic health records (observations)
• pre-clinical associations:
– genotype-phenotype (null/disease models) [r2:mgi, r2:sgd; r3:wormbase]
– in vitro assays (IC50) [r3:chembl]
– drug targets [r2:drugbank; r2:ctd; r3:stitch]
– drug-gene expression [r3:gxa]
– pathways [r2:kegg; r3:reactome]
– Drug-pathway, disease-pathway enrichments [aberrant pathways]
– Chemical properties [r2:pubchem; r2.drugbank]
– Toxicology [r1.toxkb/cebs]
@micheldumontier::ACS:23-08-1620](https://image.slidesharecdn.com/2016-acs-skolnik-award-symposium-dumontier-160823233539/75/Building-a-Network-of-Interoperable-and-Independently-Produced-Linked-and-Open-Biomedical-Data-20-2048.jpg)