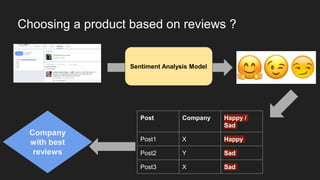
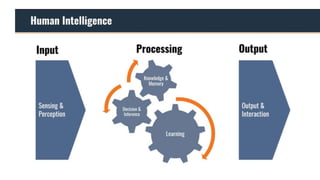
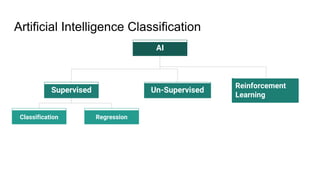
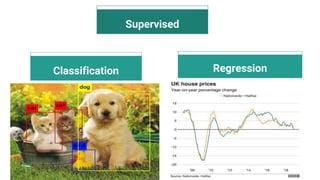
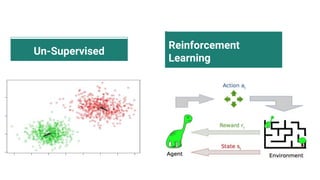
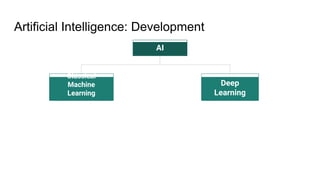
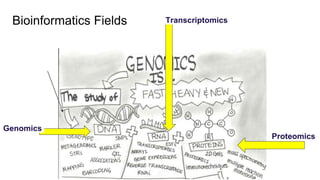
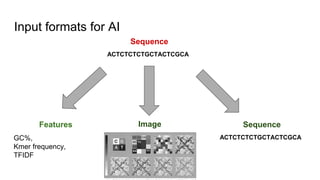
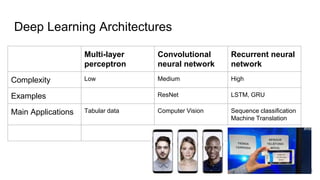
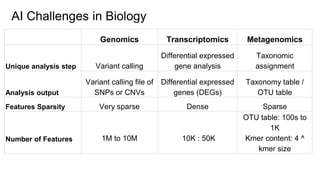
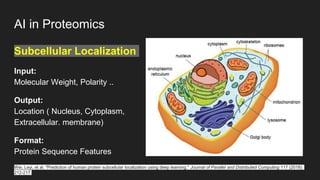
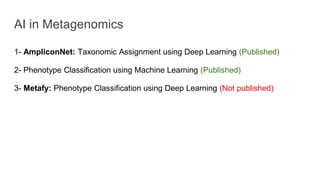
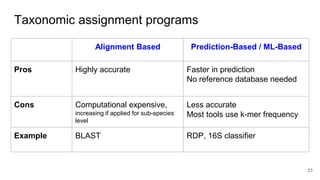
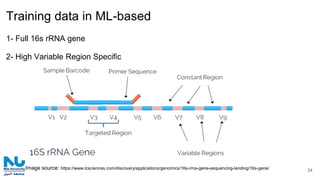
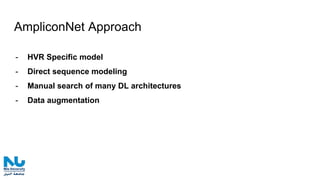
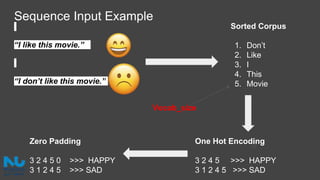
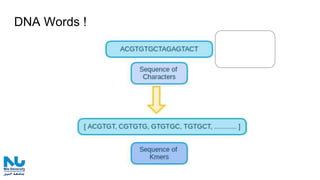
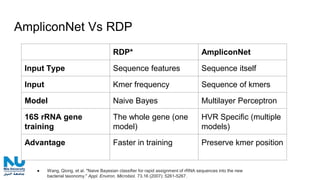
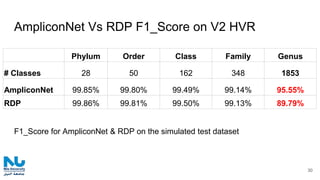
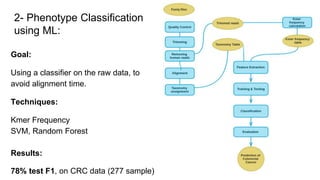
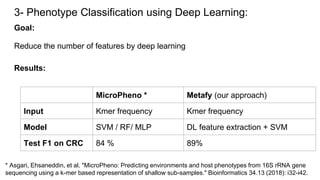
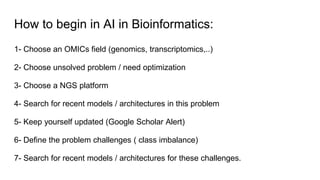
The document discusses the application of artificial intelligence (AI) and deep learning in bioinformatics, specifically focusing on areas such as genomics, transcriptomics, and metagenomics. It outlines various AI challenges, models, and methods used for tasks like sentiment analysis, taxonomic classification, and phenotype classification. The document also provides insights into how to start implementing AI in bioinformatics and emphasizes the importance of understanding general AI principles before delving into specific problems.