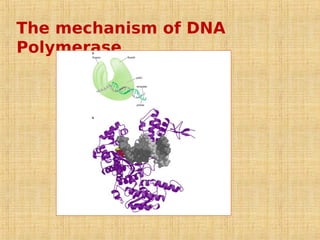
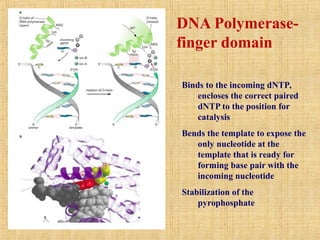
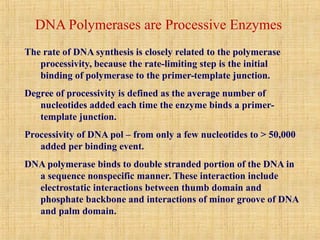
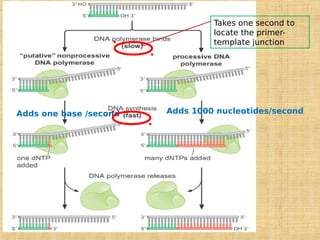
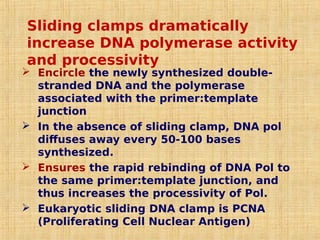
1. DNA replication enzymes and mechanisms ensure accurate and complete duplication of the genome during cell division.
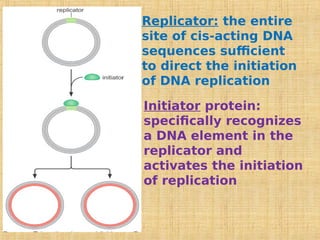
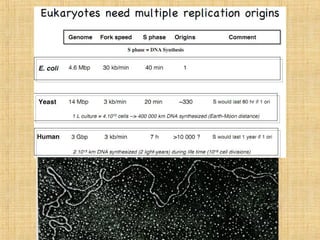
2. Specific DNA sequences called origins of replication initiate DNA unwinding and replication at replication forks.
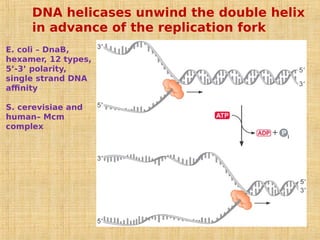
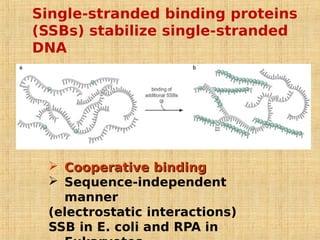
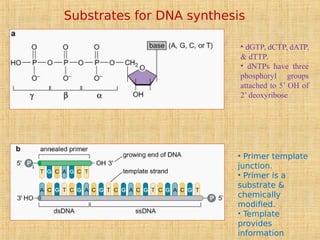
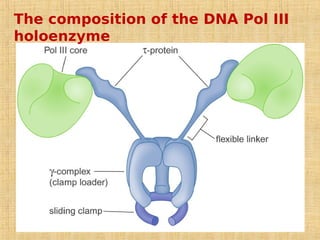
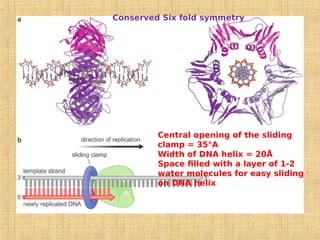
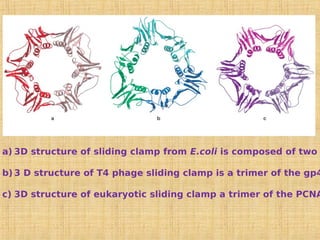
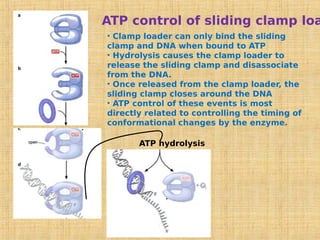
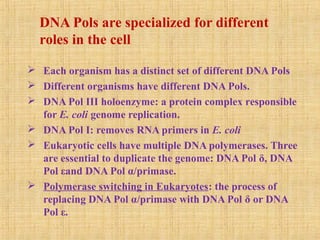
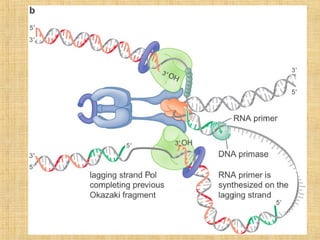
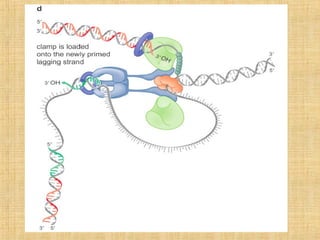
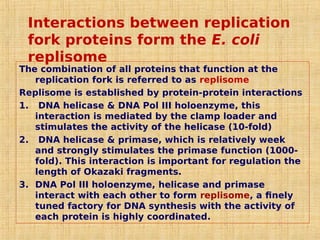
3. Replication enzymes include helicases, primases, DNA polymerases, topoisomerases, ligases and sliding clamps that work together in the replisome complex.
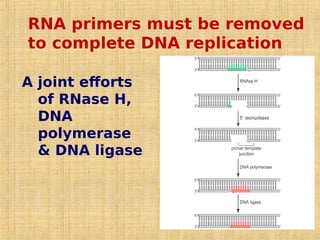
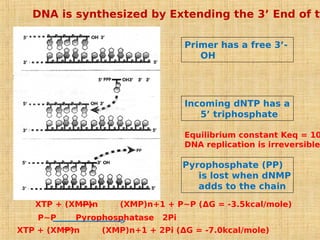
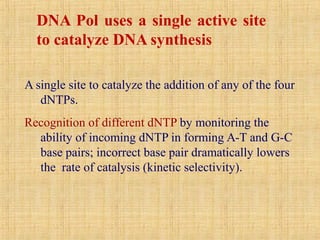
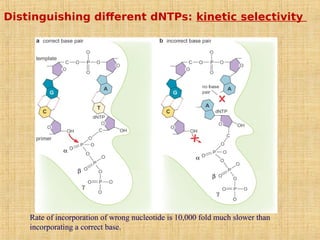
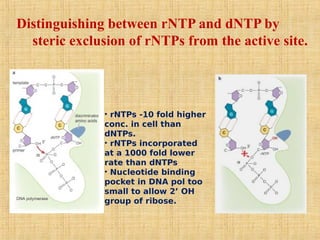
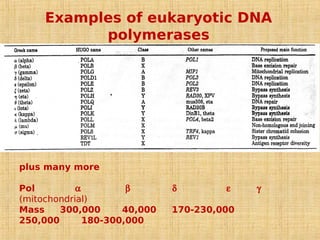
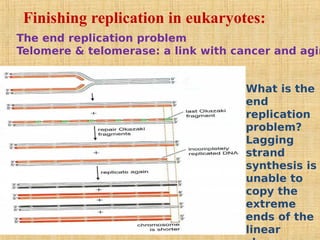
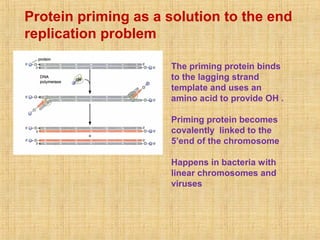
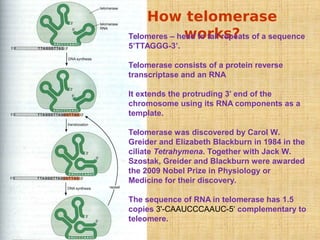
4. Elaborate mechanisms such as proofreading and processive polymerases minimize errors during DNA synthesis.