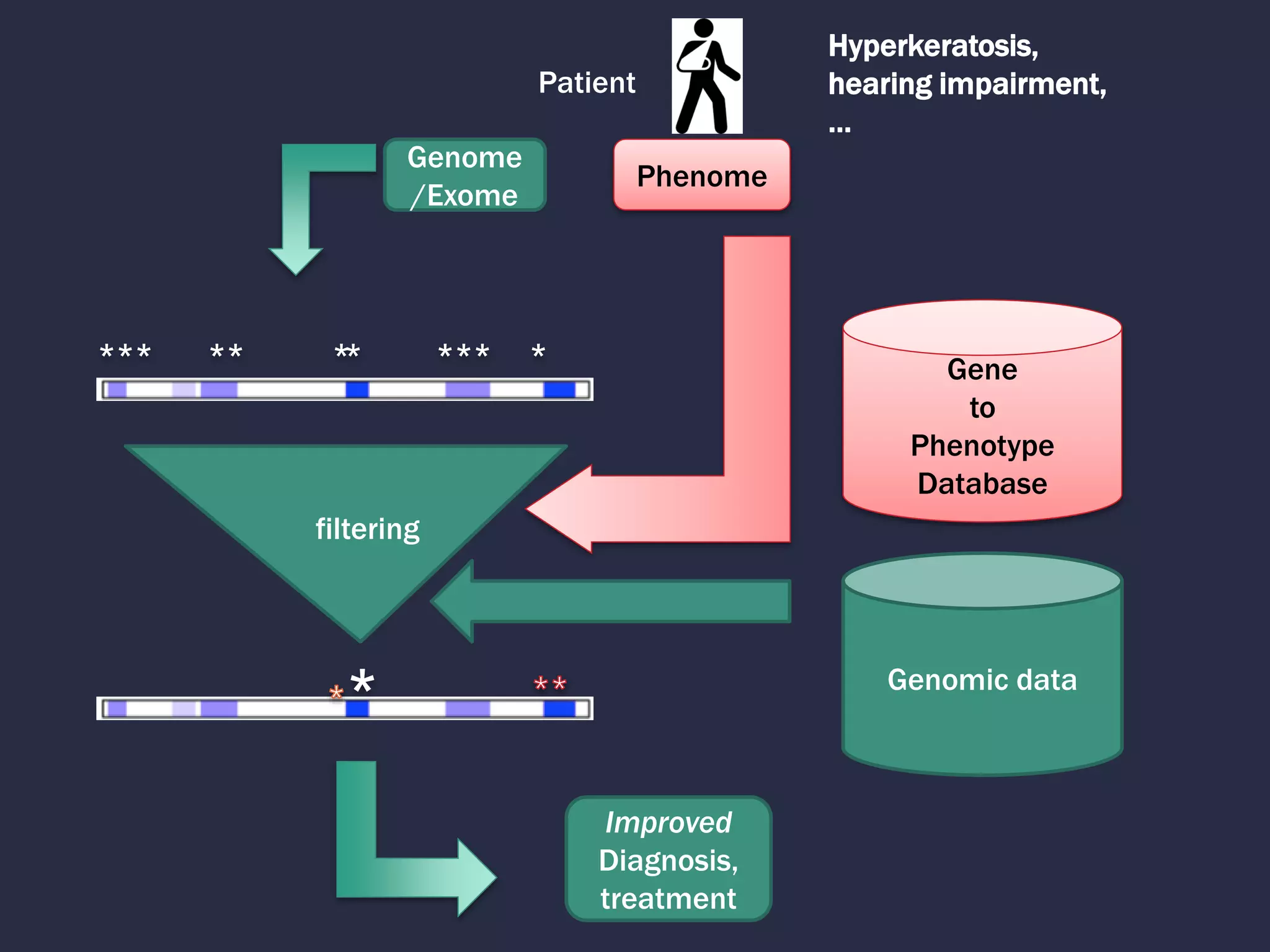
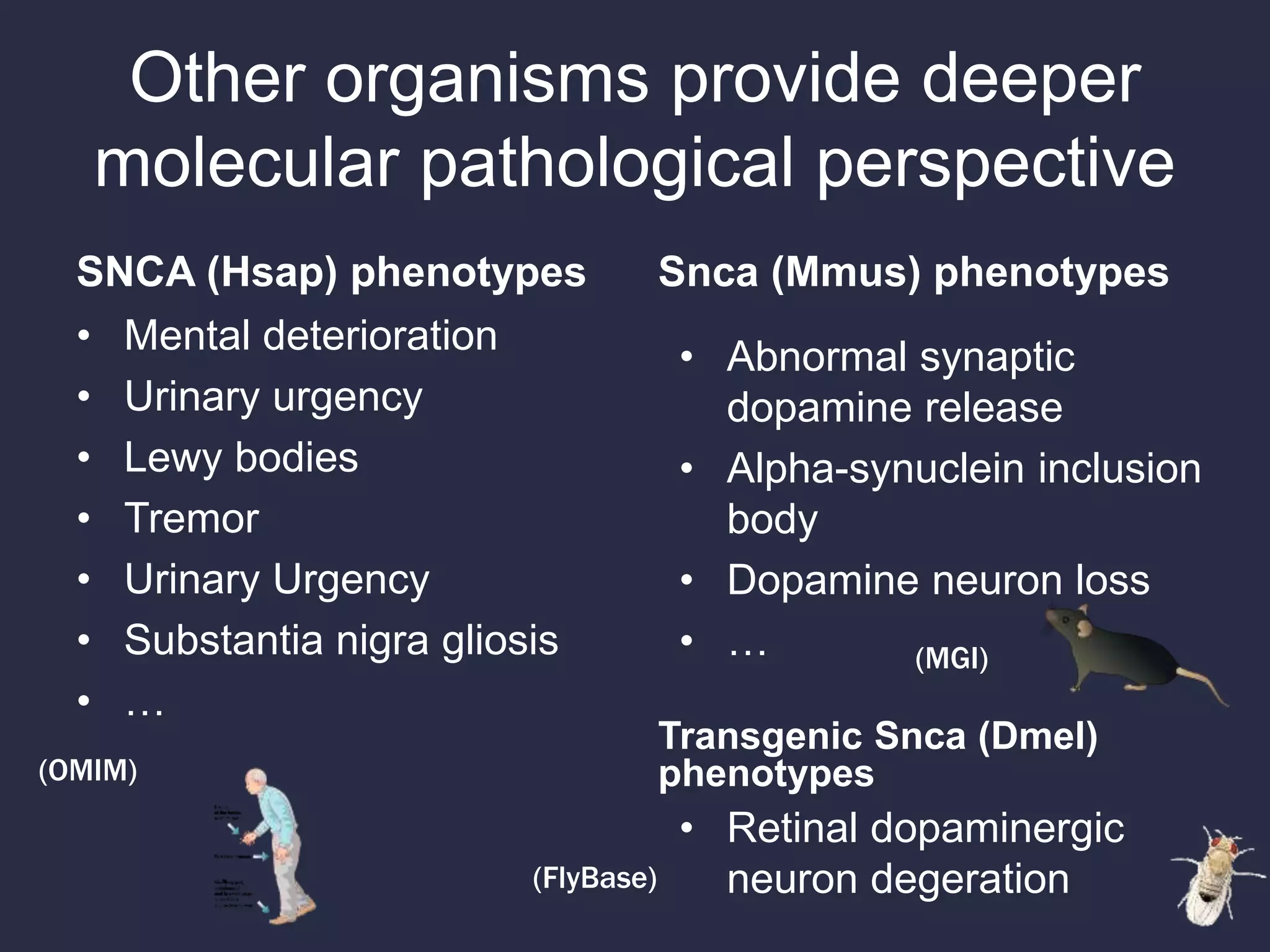
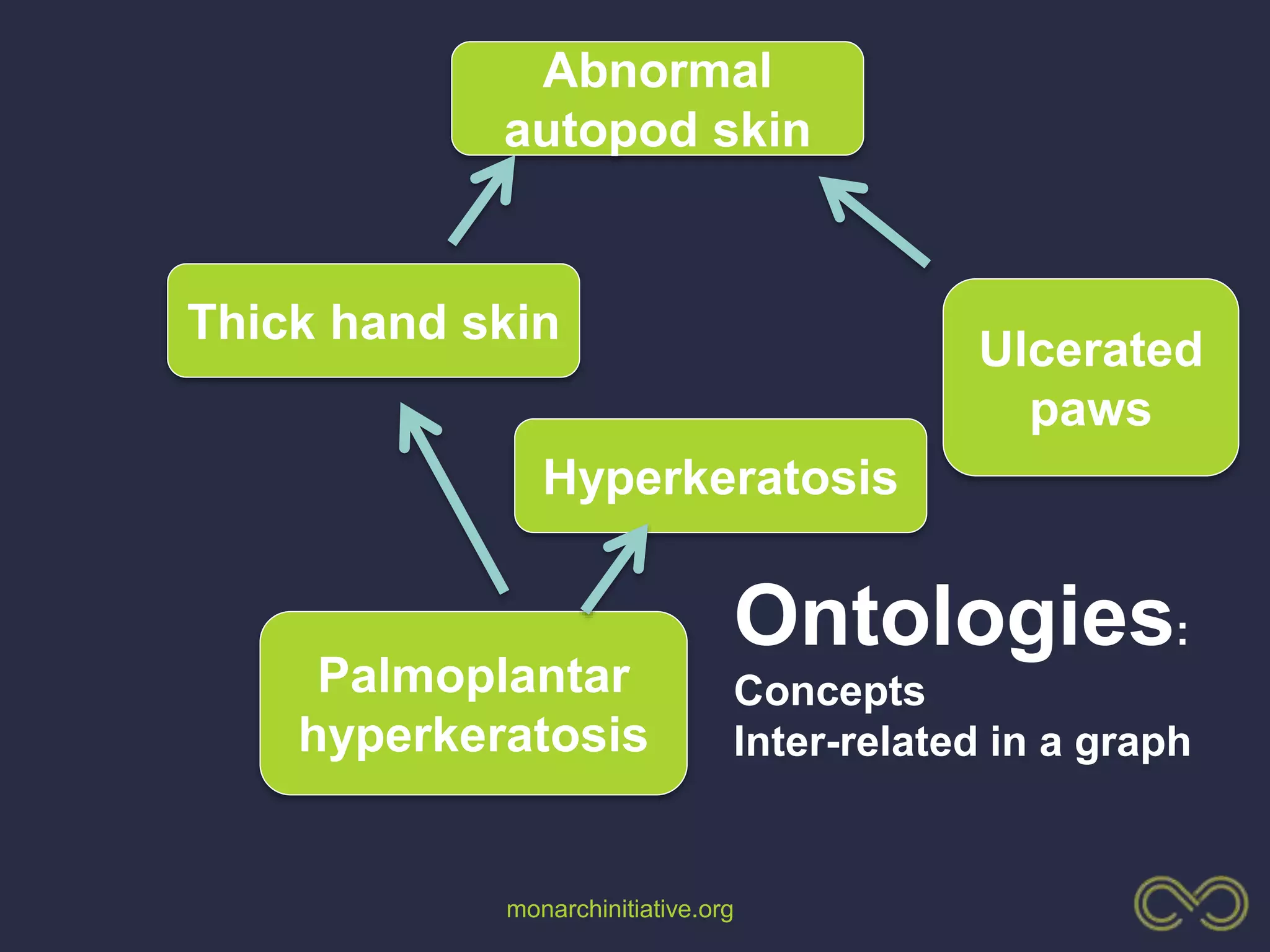
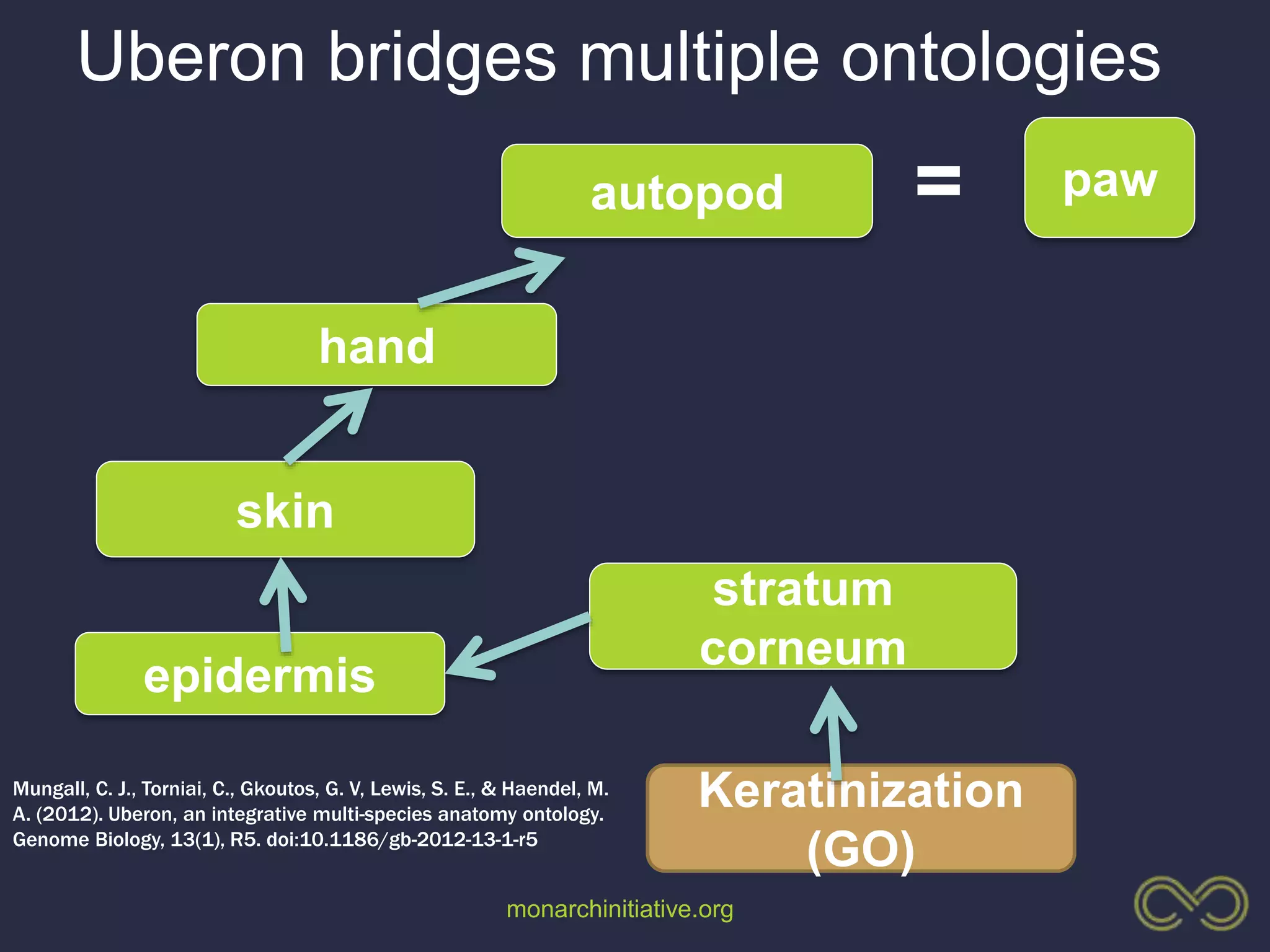
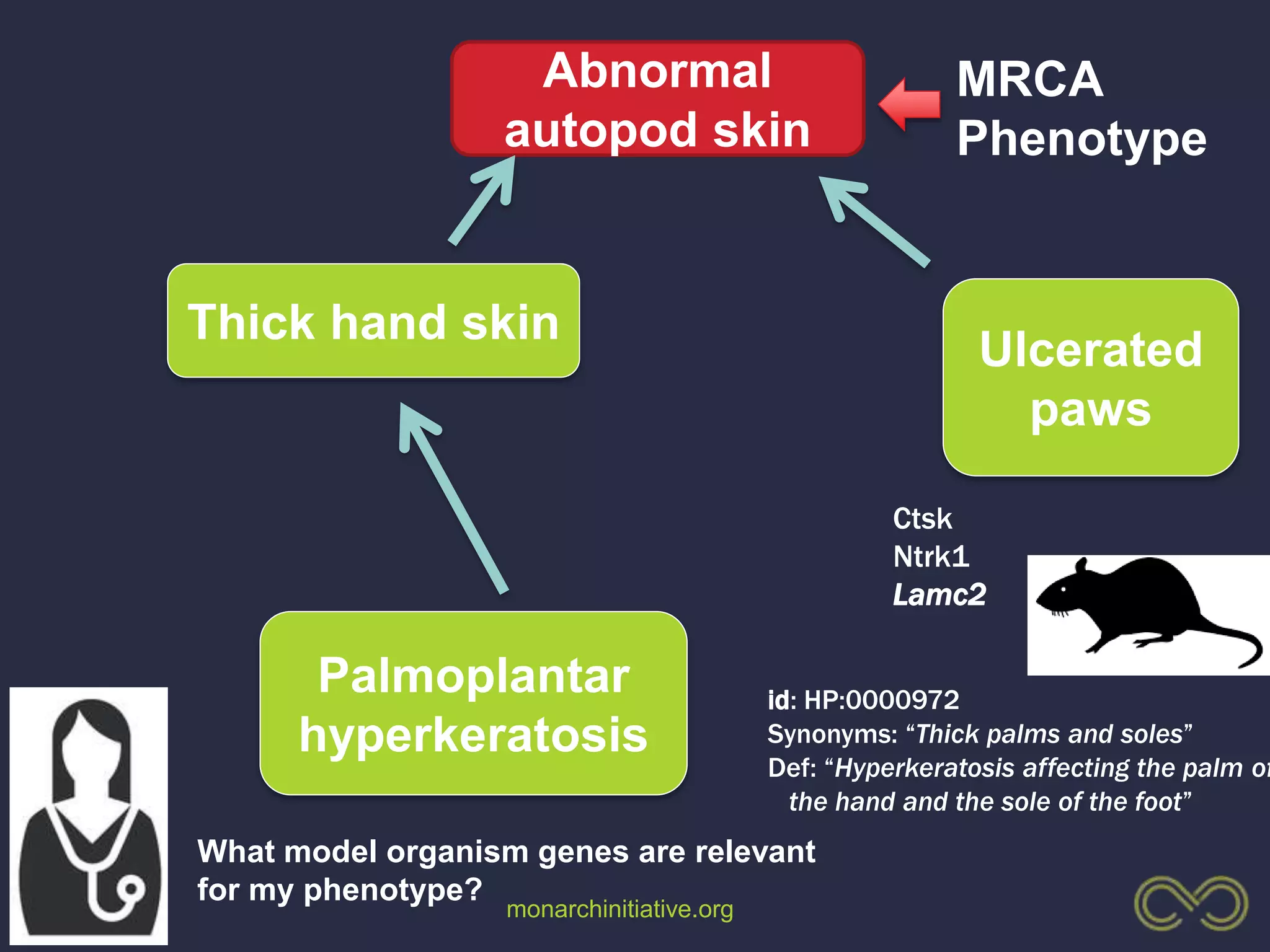
1. The document discusses using phenotypes across species to aid in interpreting genomic data from patients and improving diagnosis and treatment.
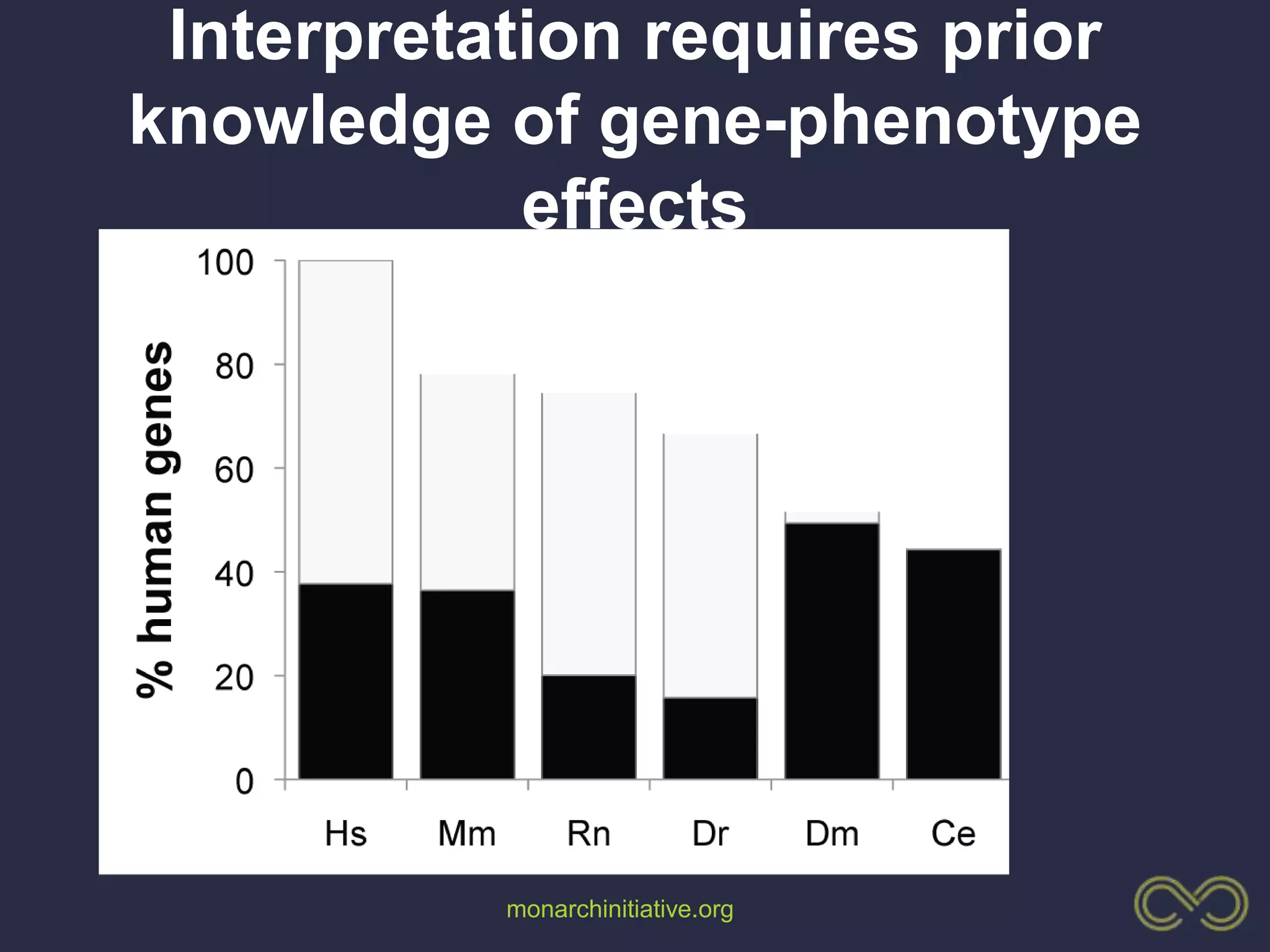
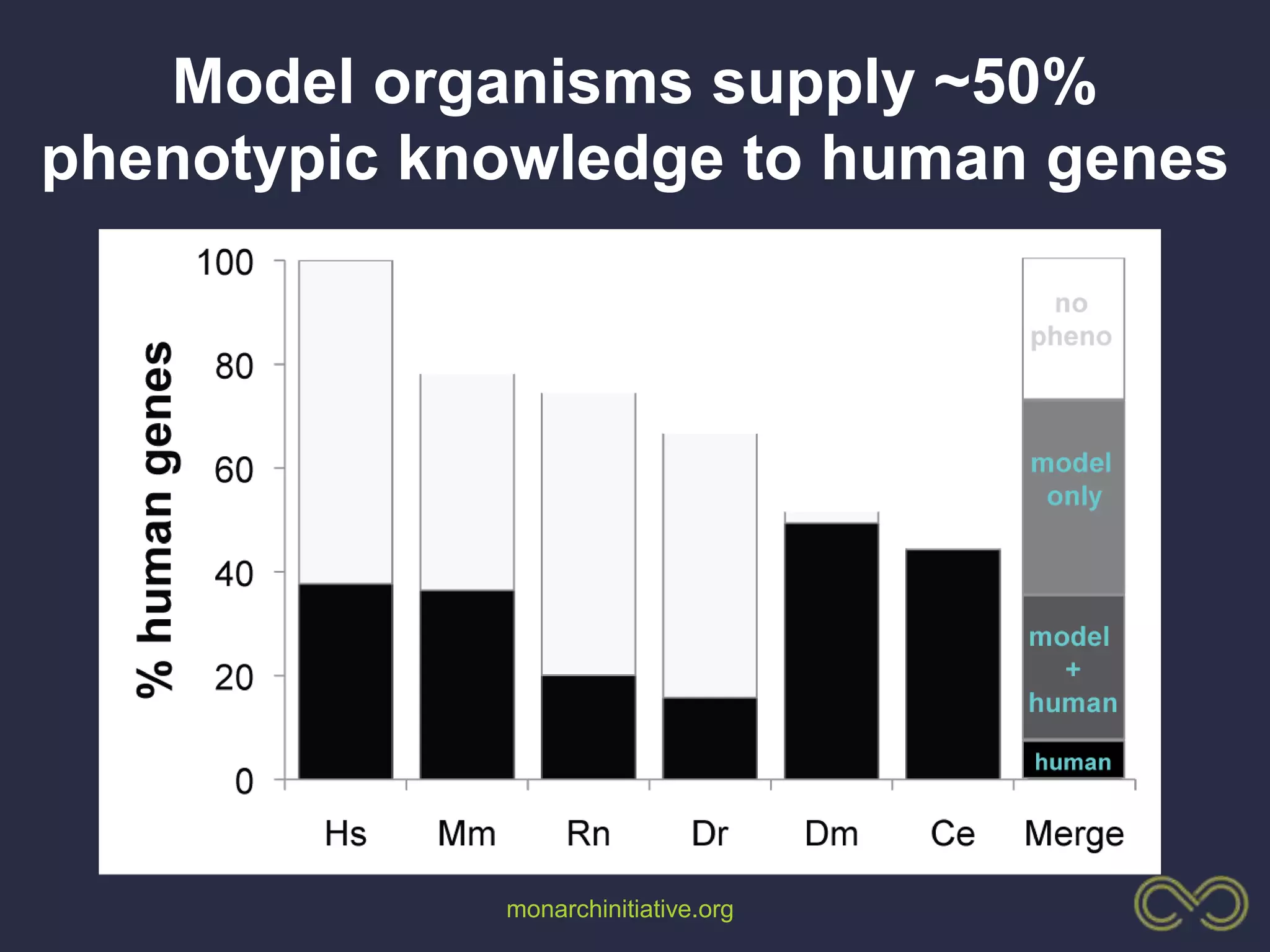
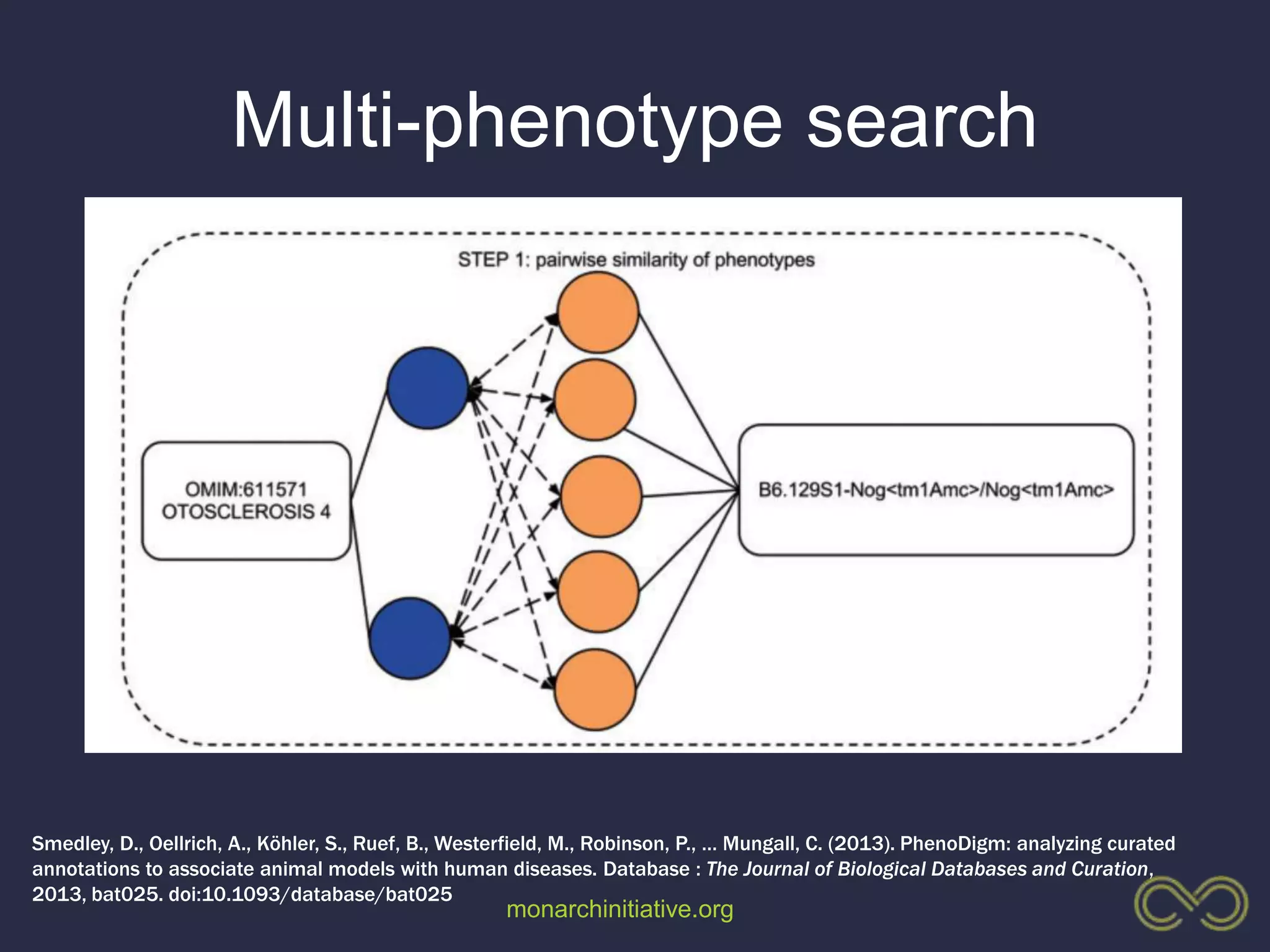
2. Building comprehensive phenotype databases from multiple sources is challenging due to disparate data on human genes/variants and model organisms.
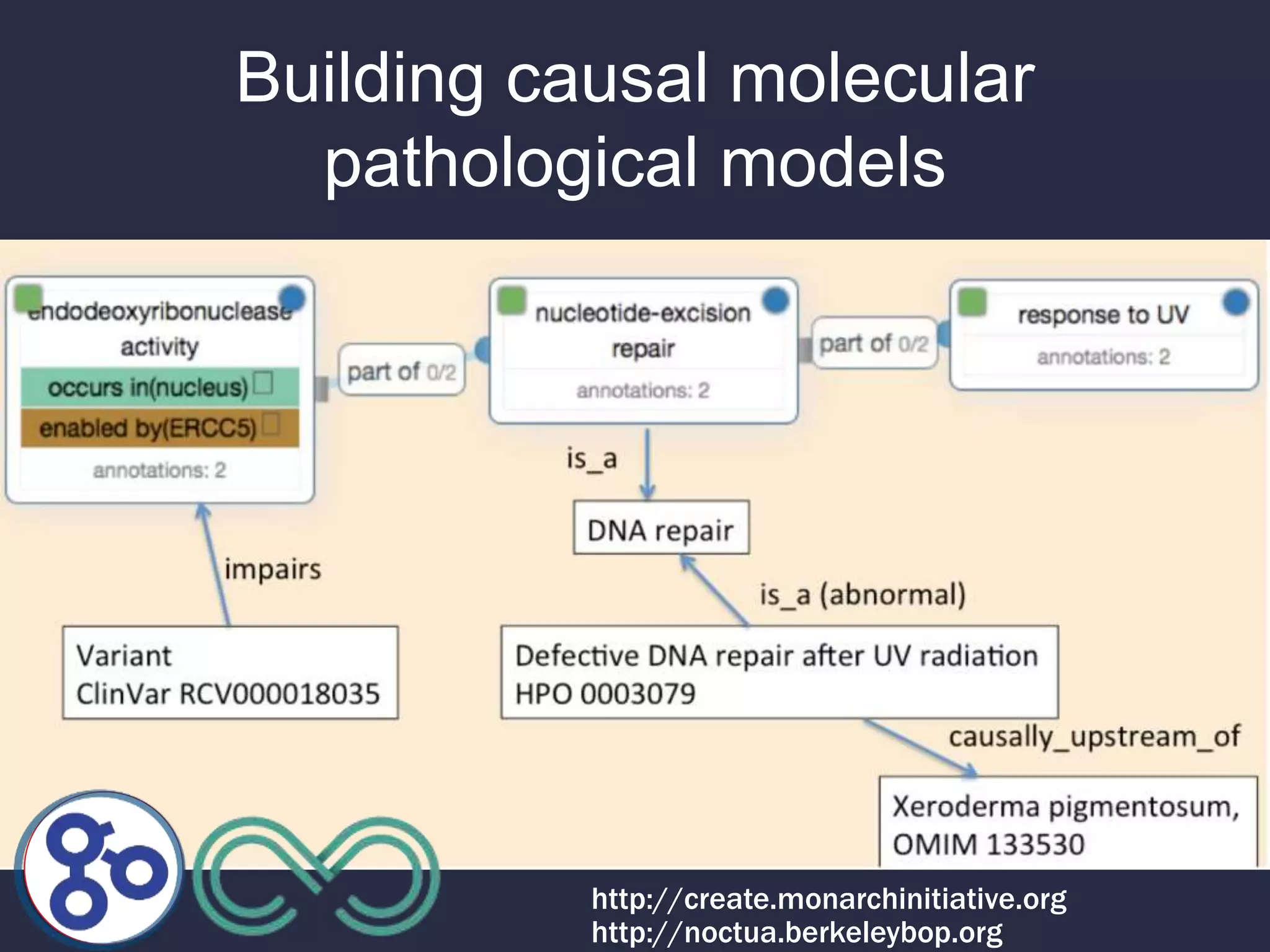
3. The Monarch Initiative aims to link human diseases to phenotypes in model systems through an ontology-based knowledge base and portal.
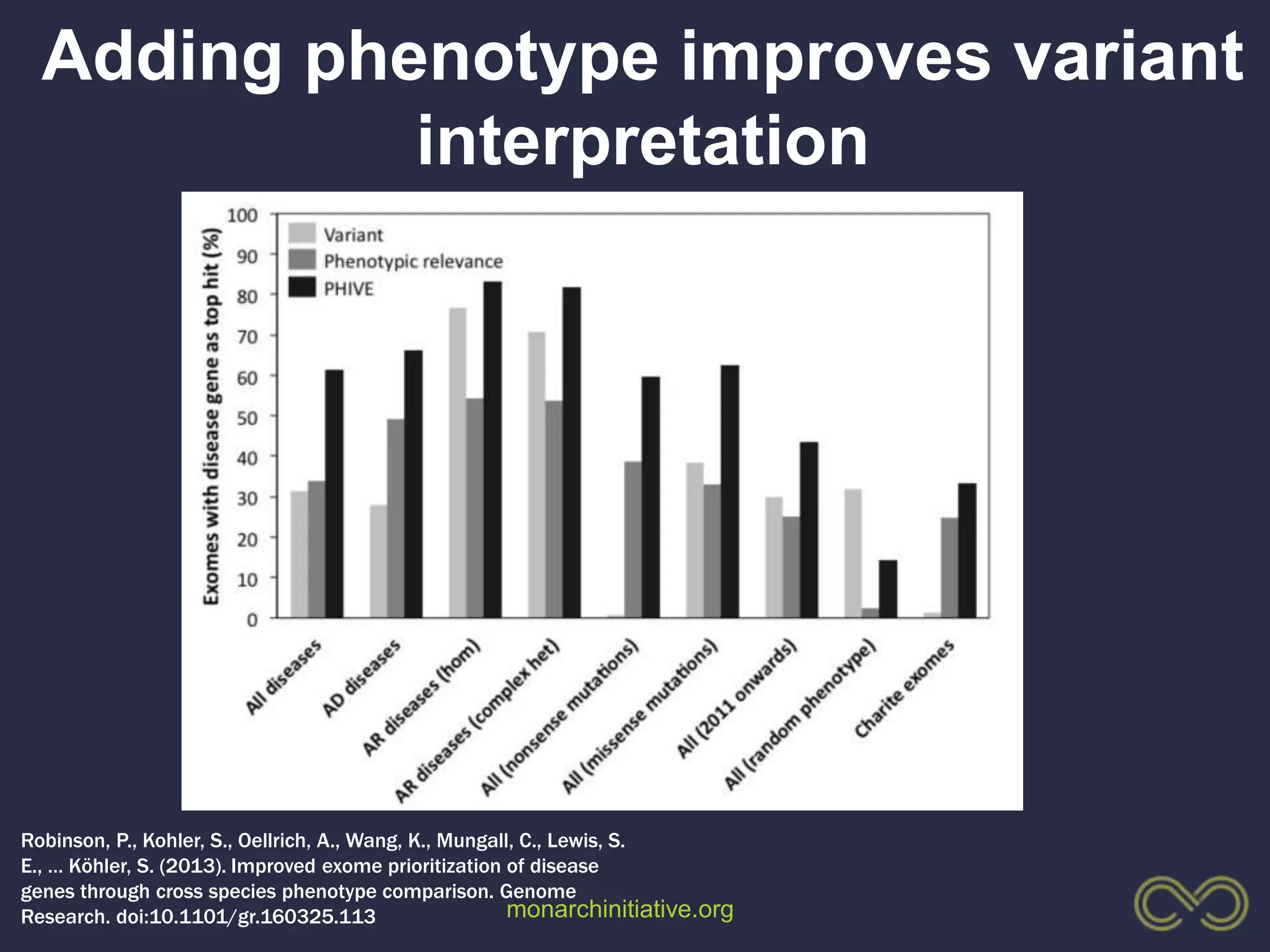
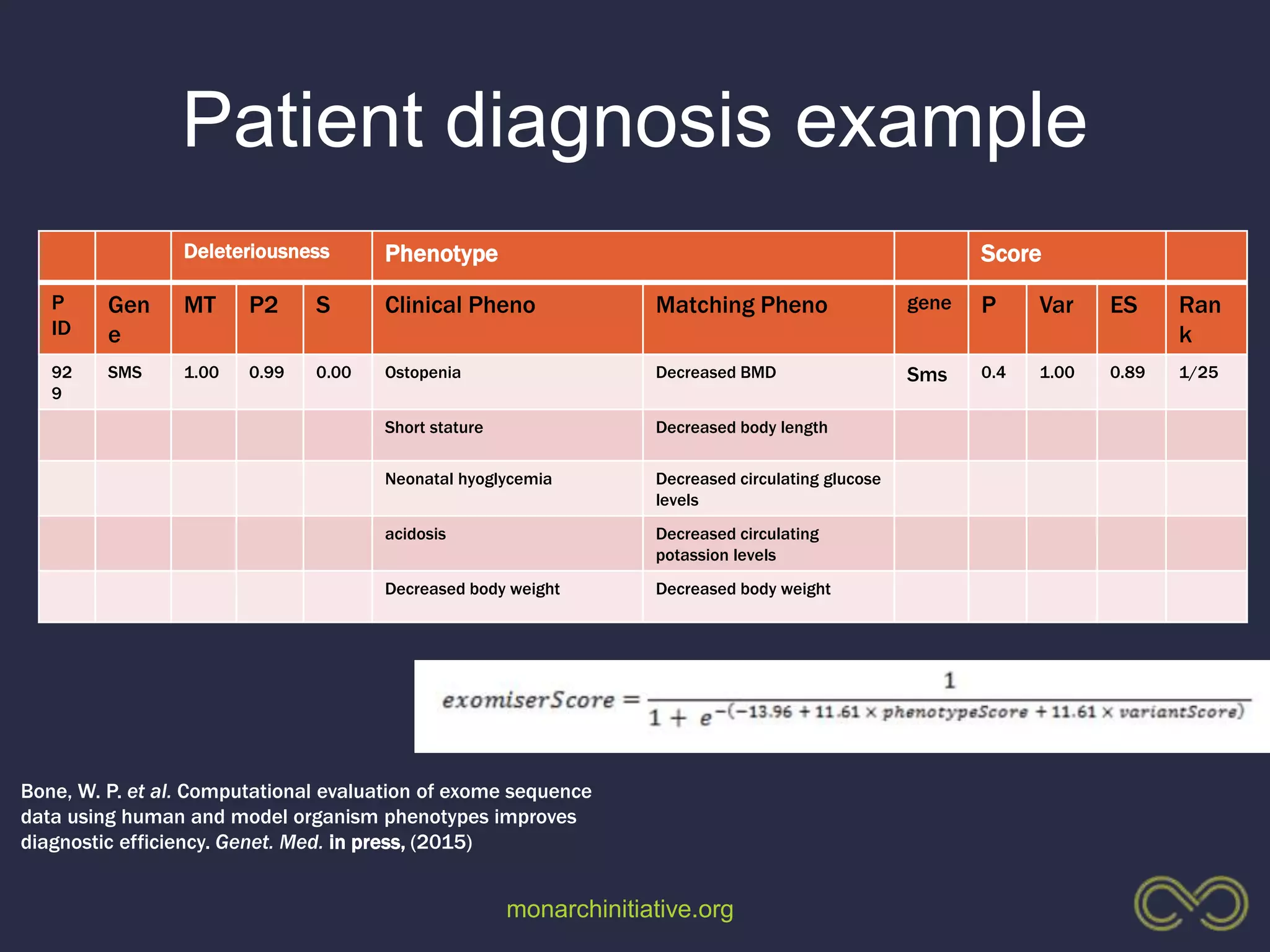
4. Incorporating rich phenotypic data can improve variant filtering and interpretation by providing more context for sequencing results.