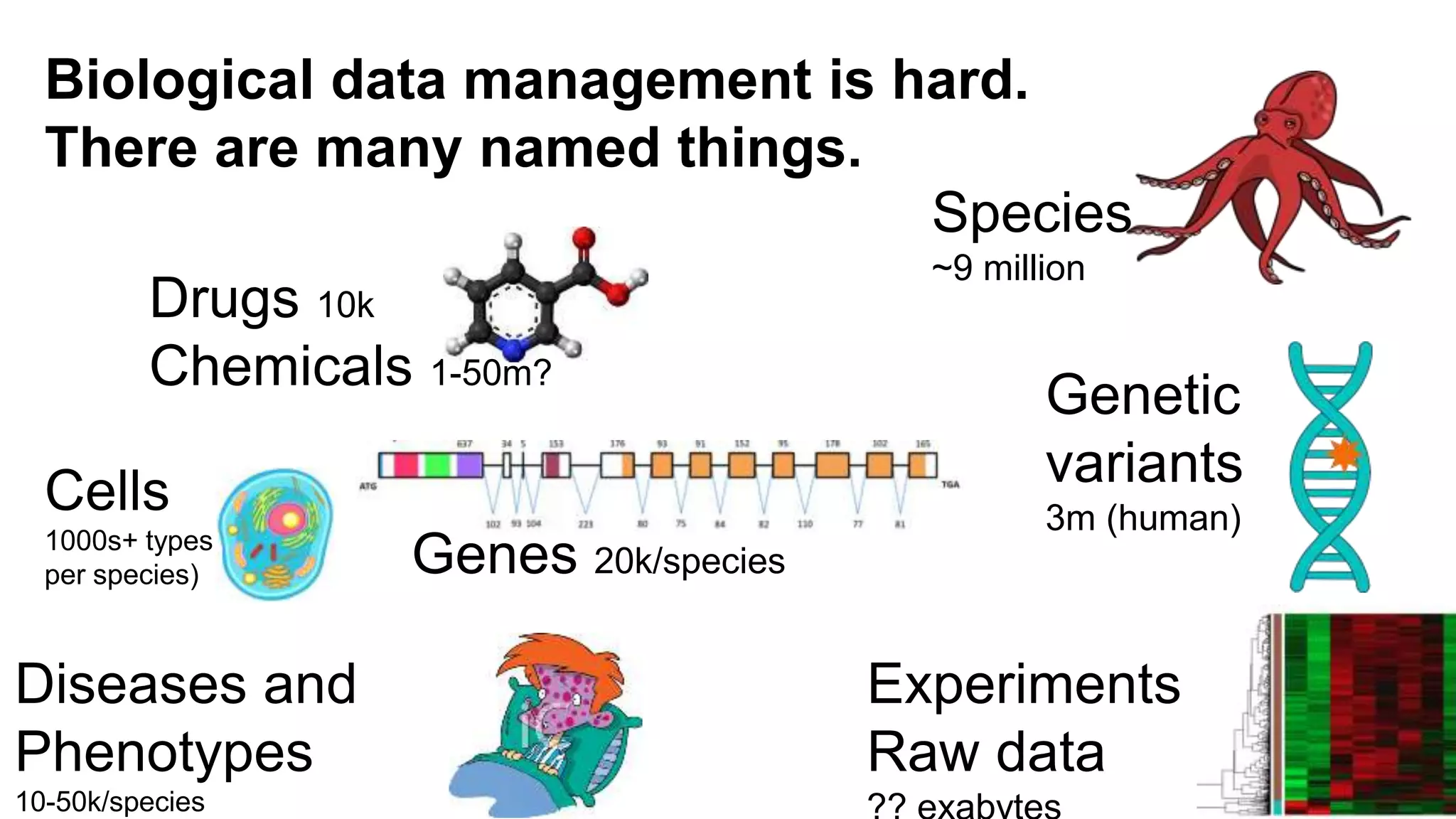
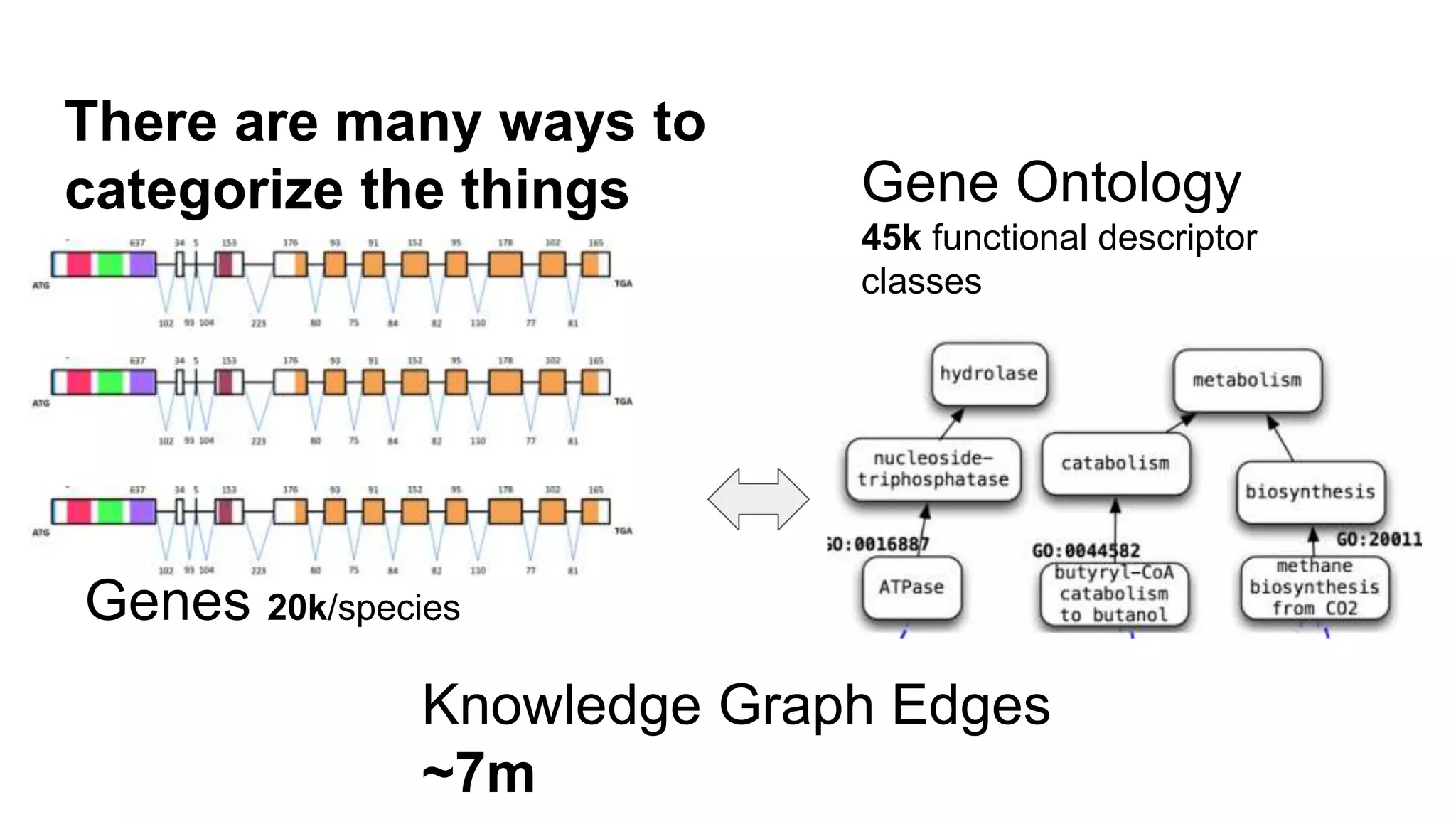
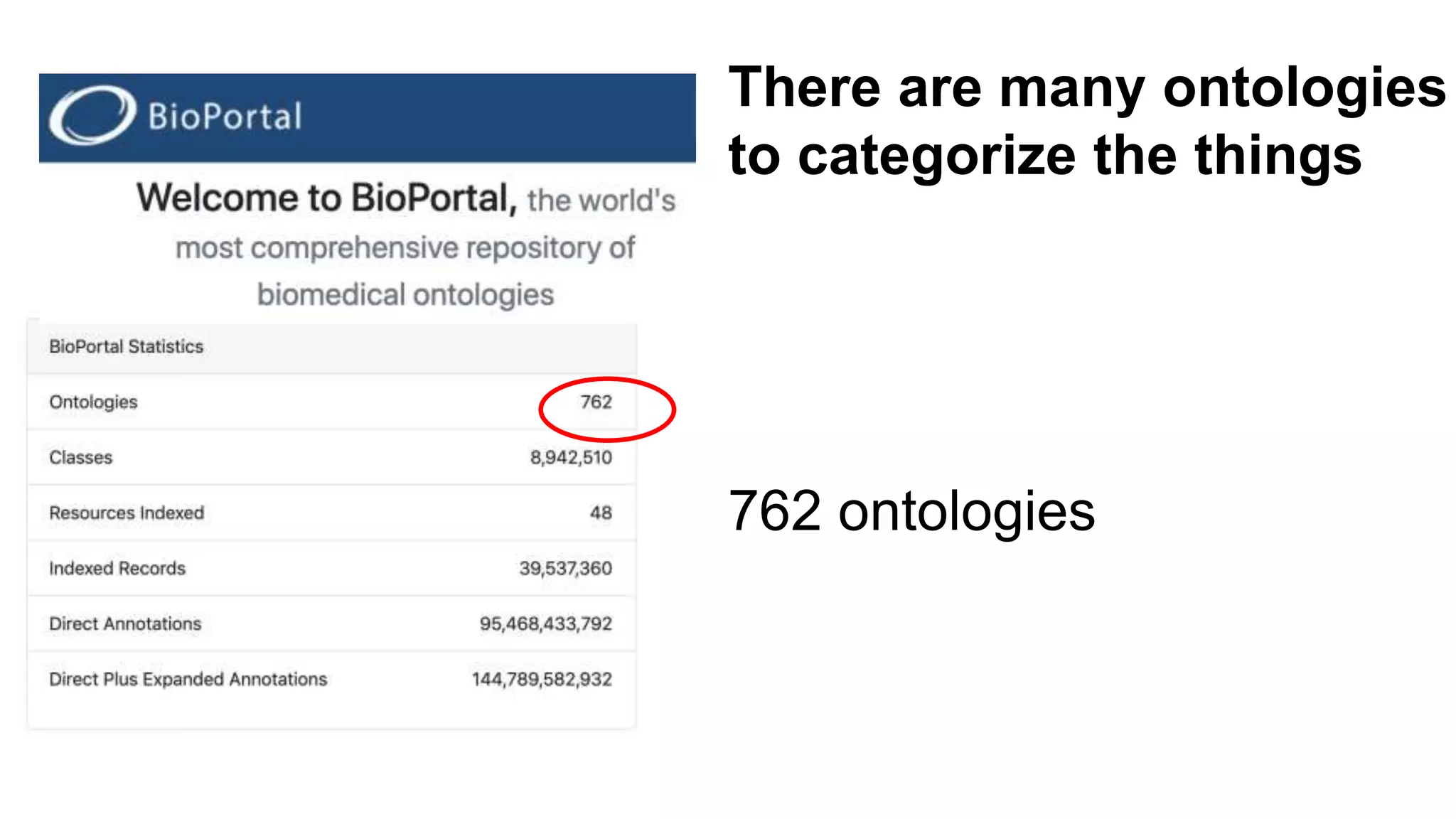
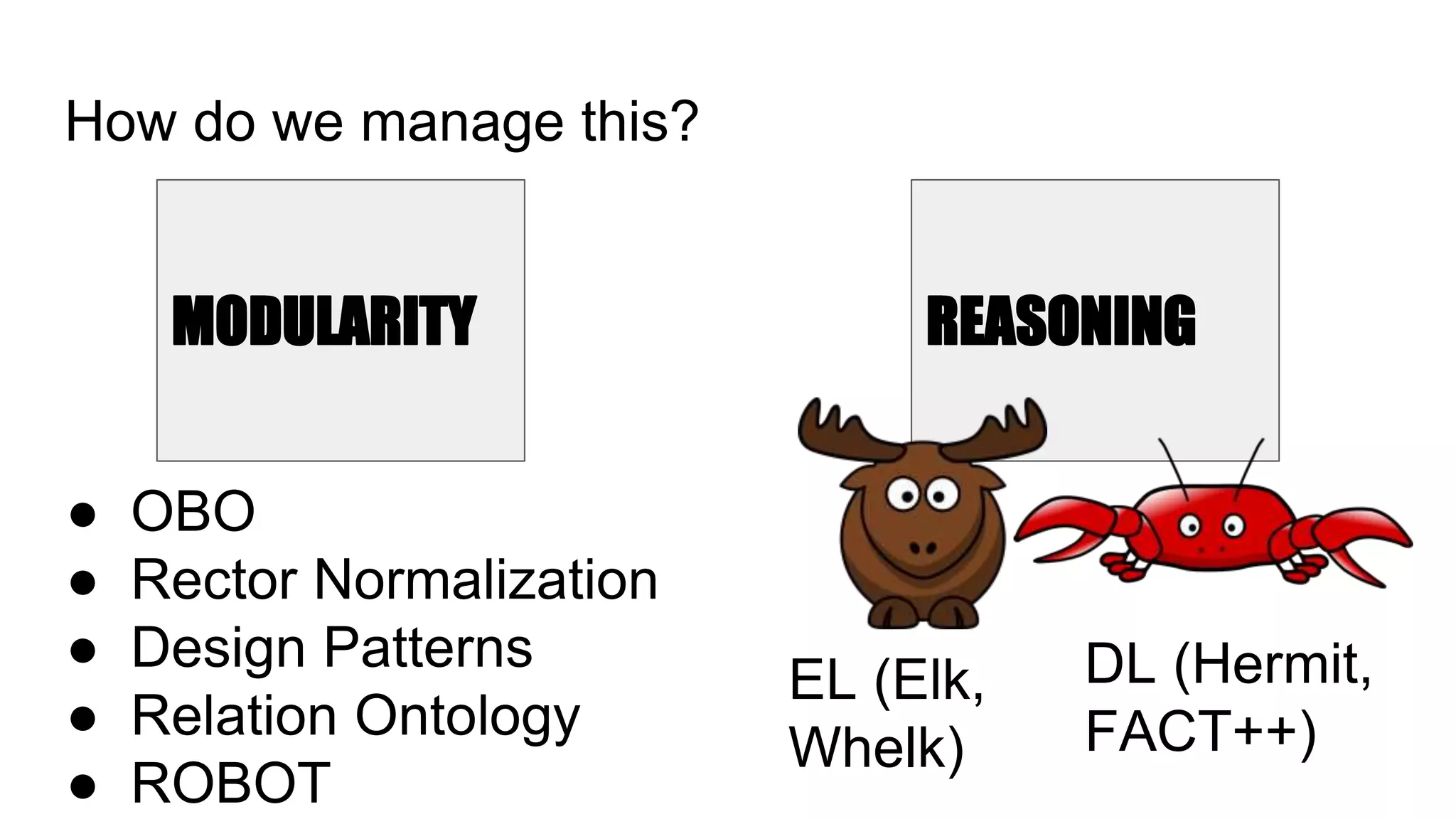
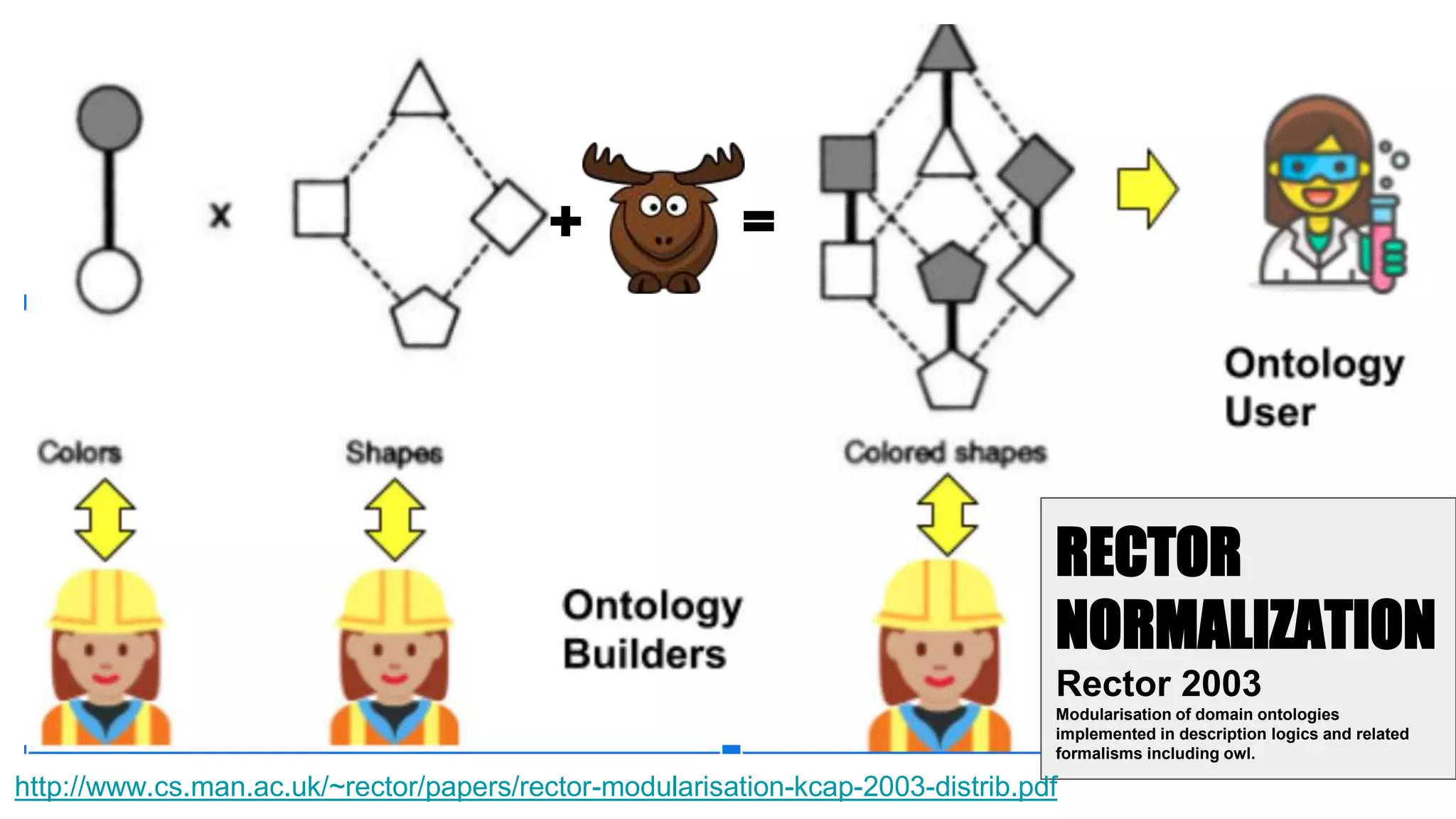
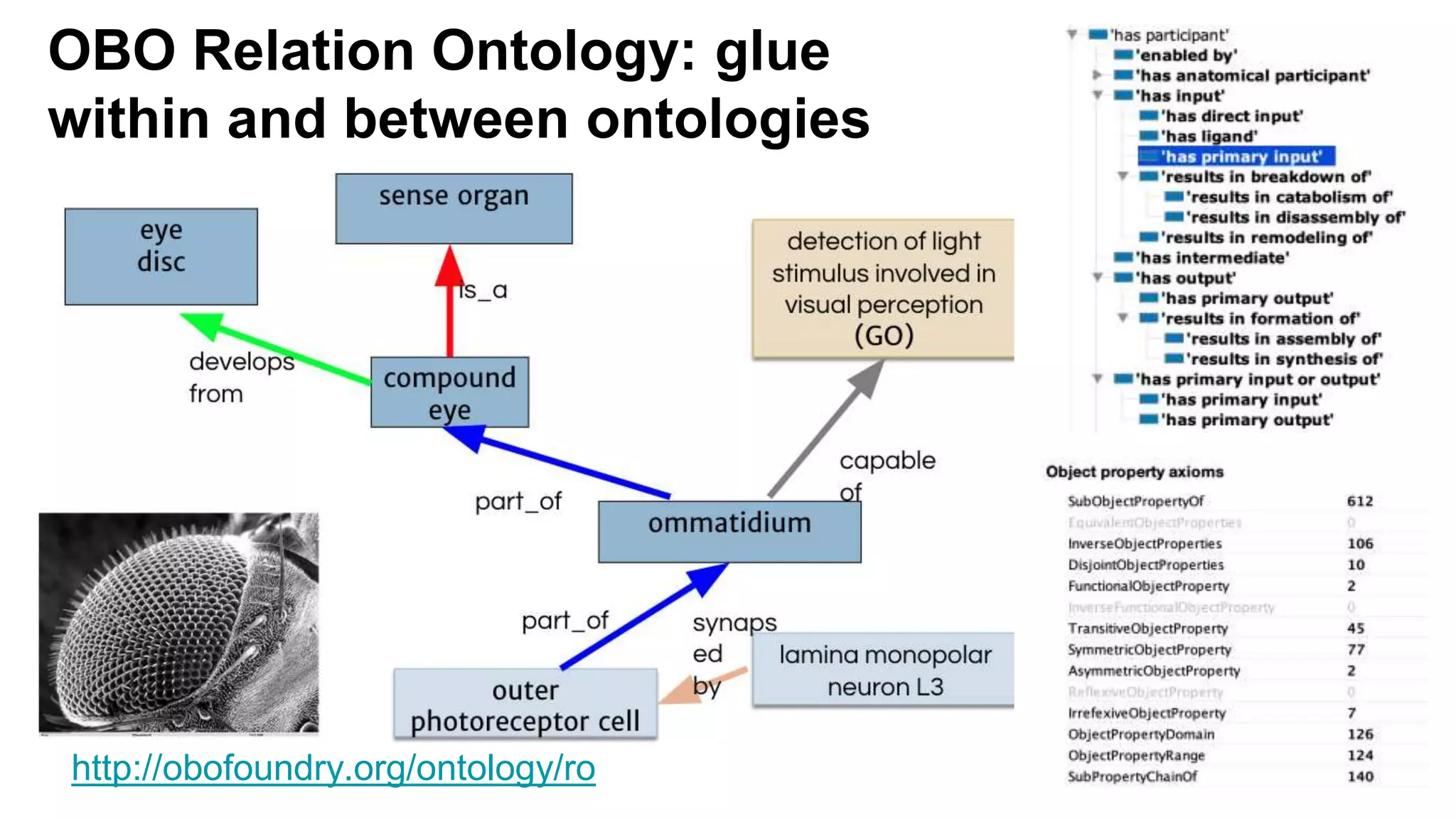
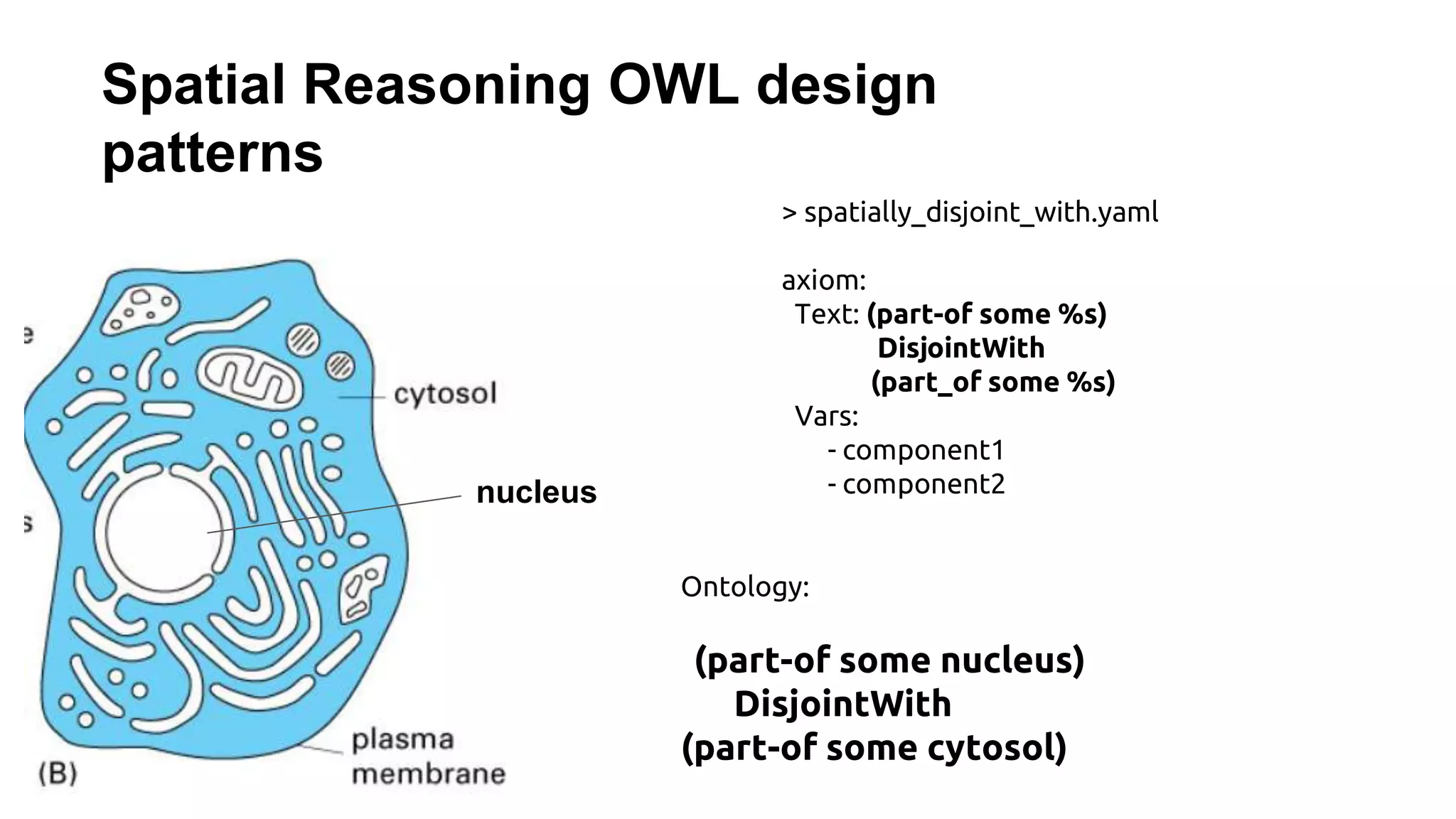
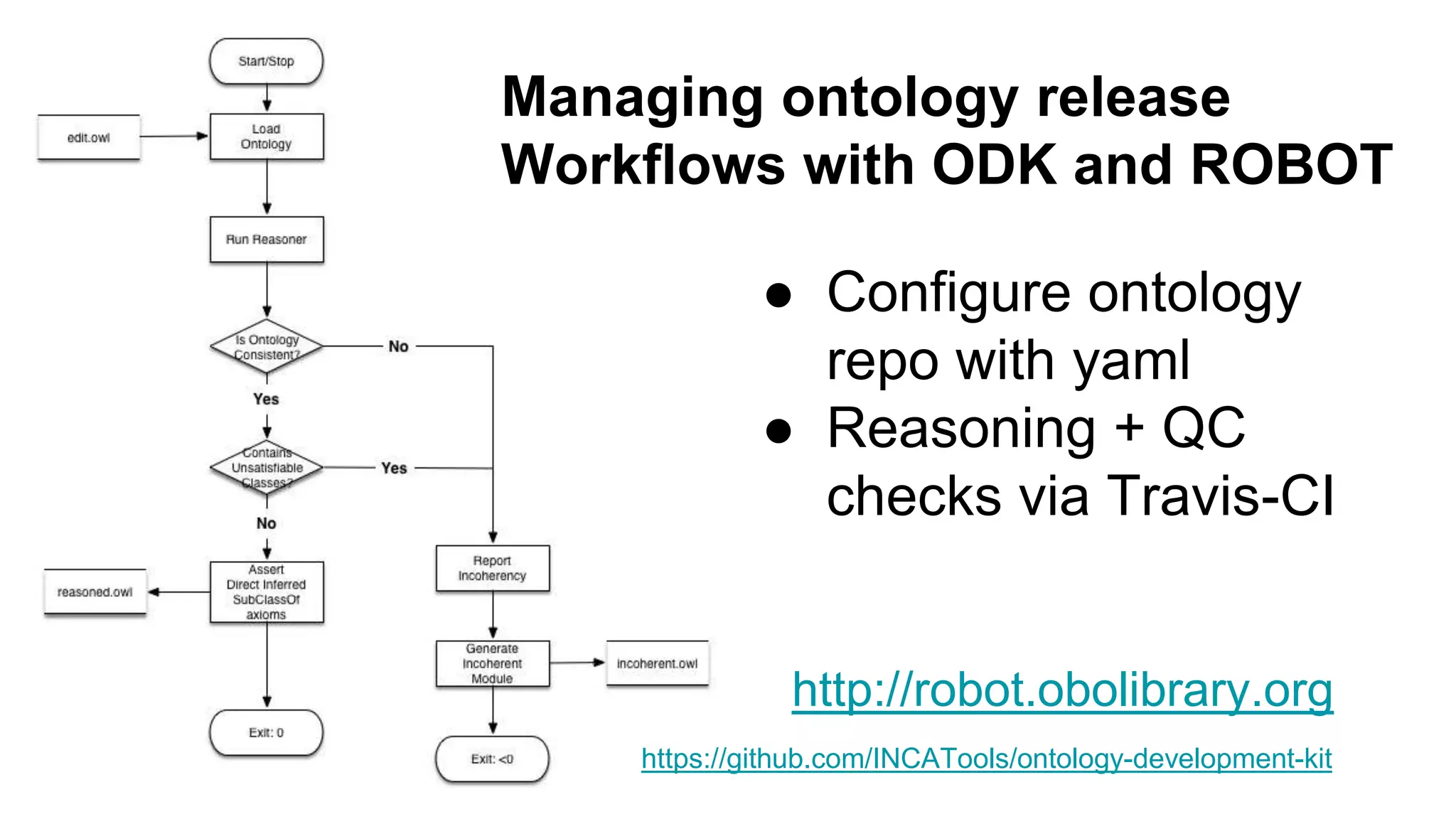
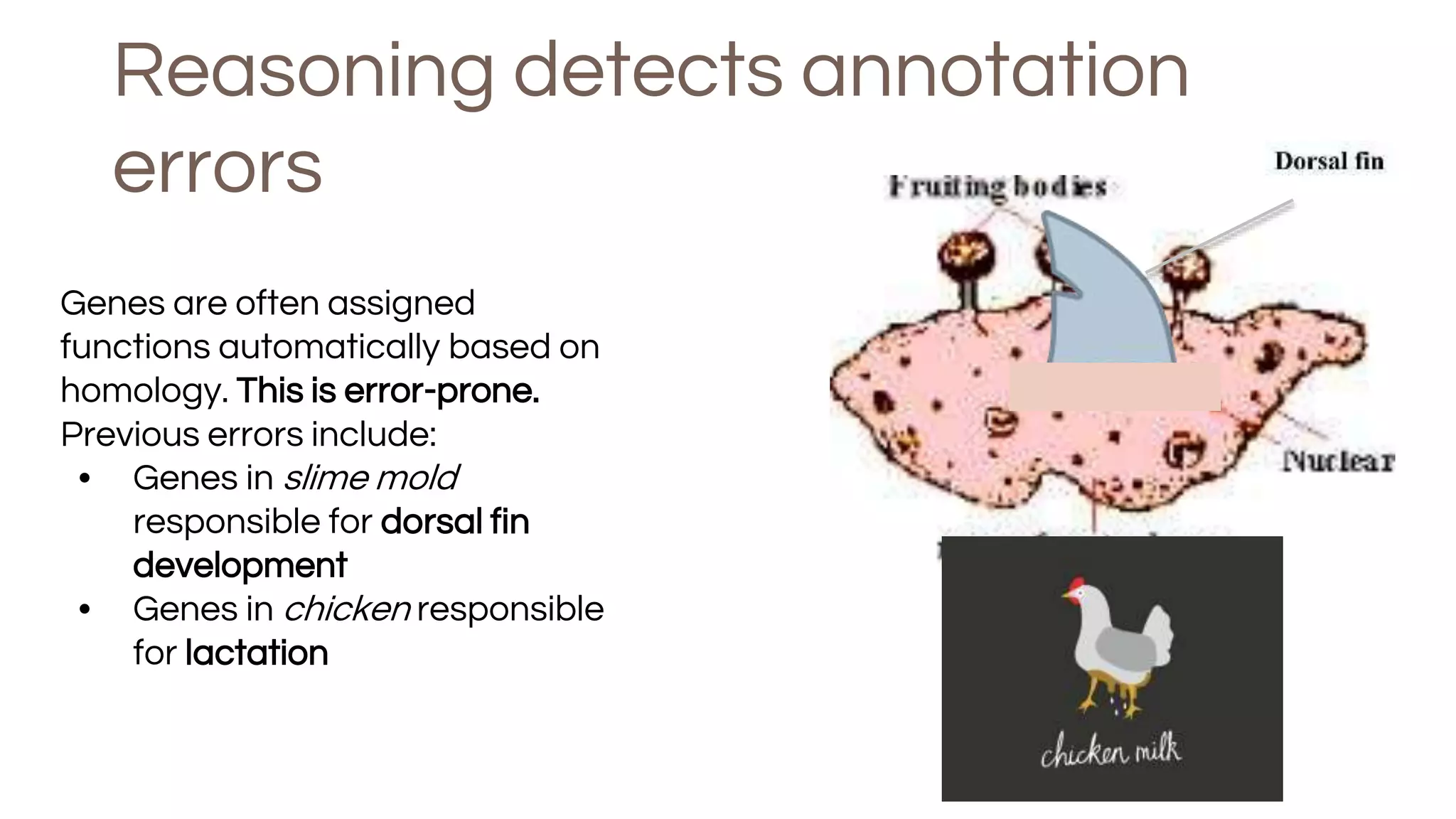
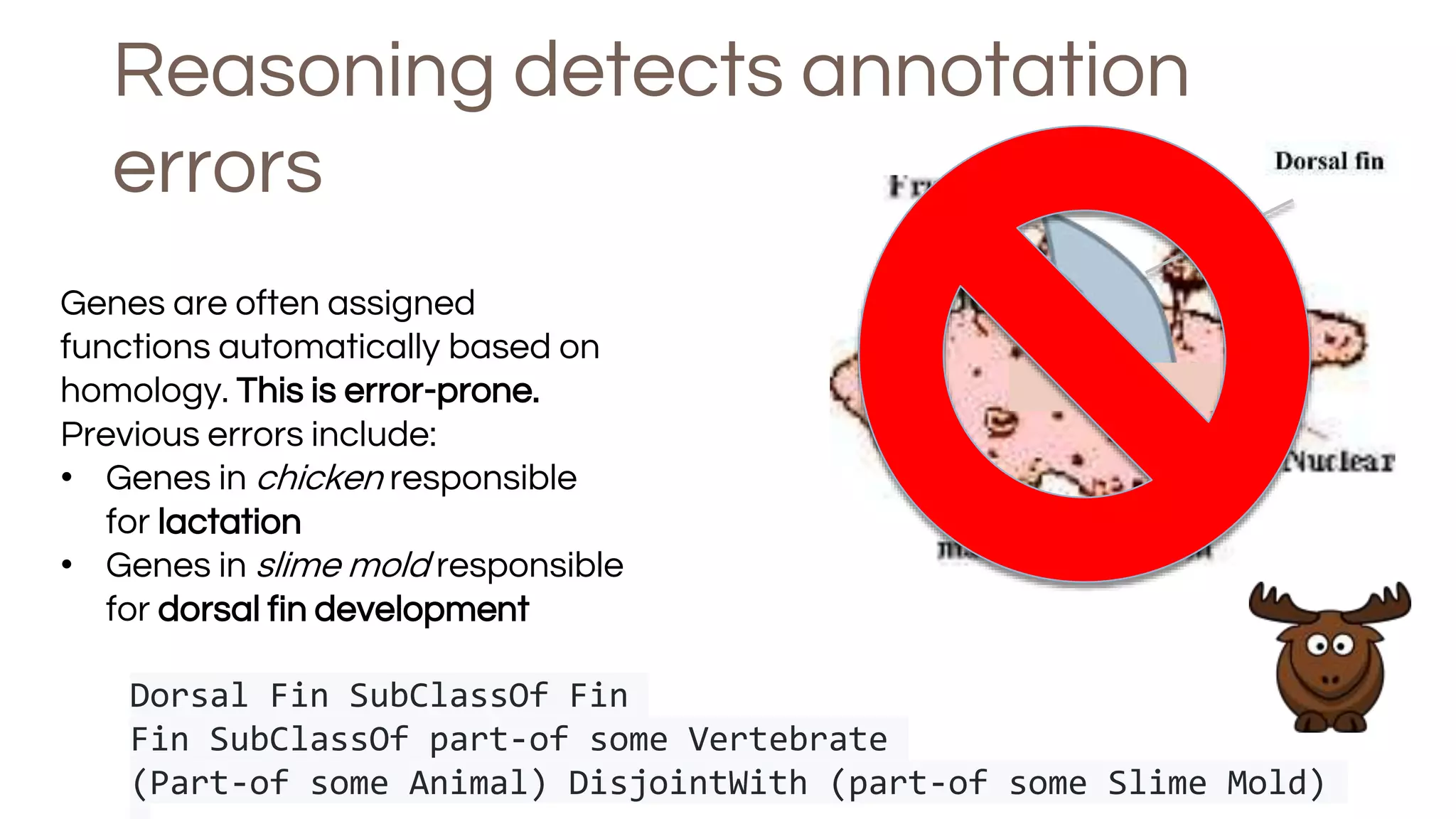
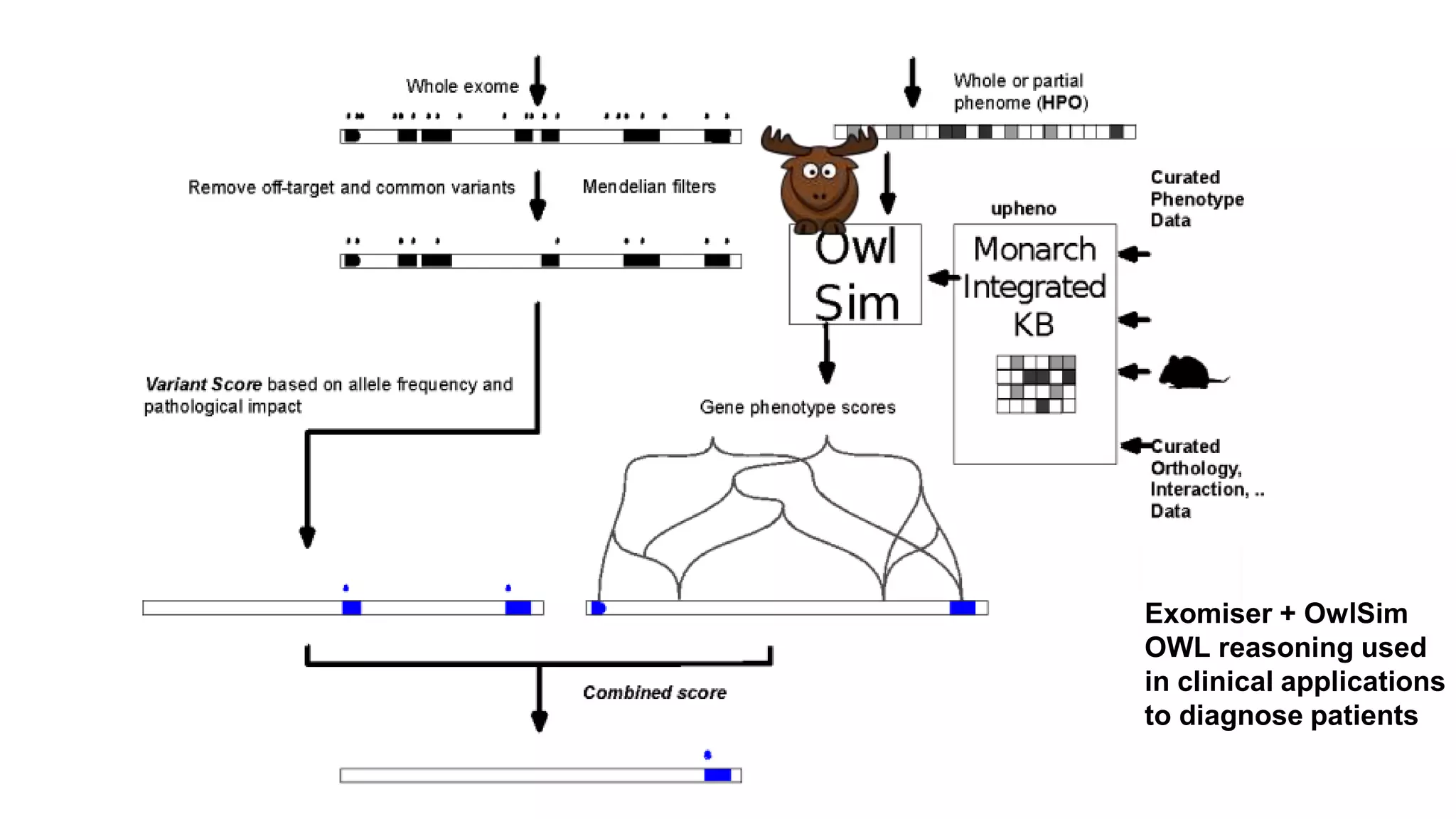
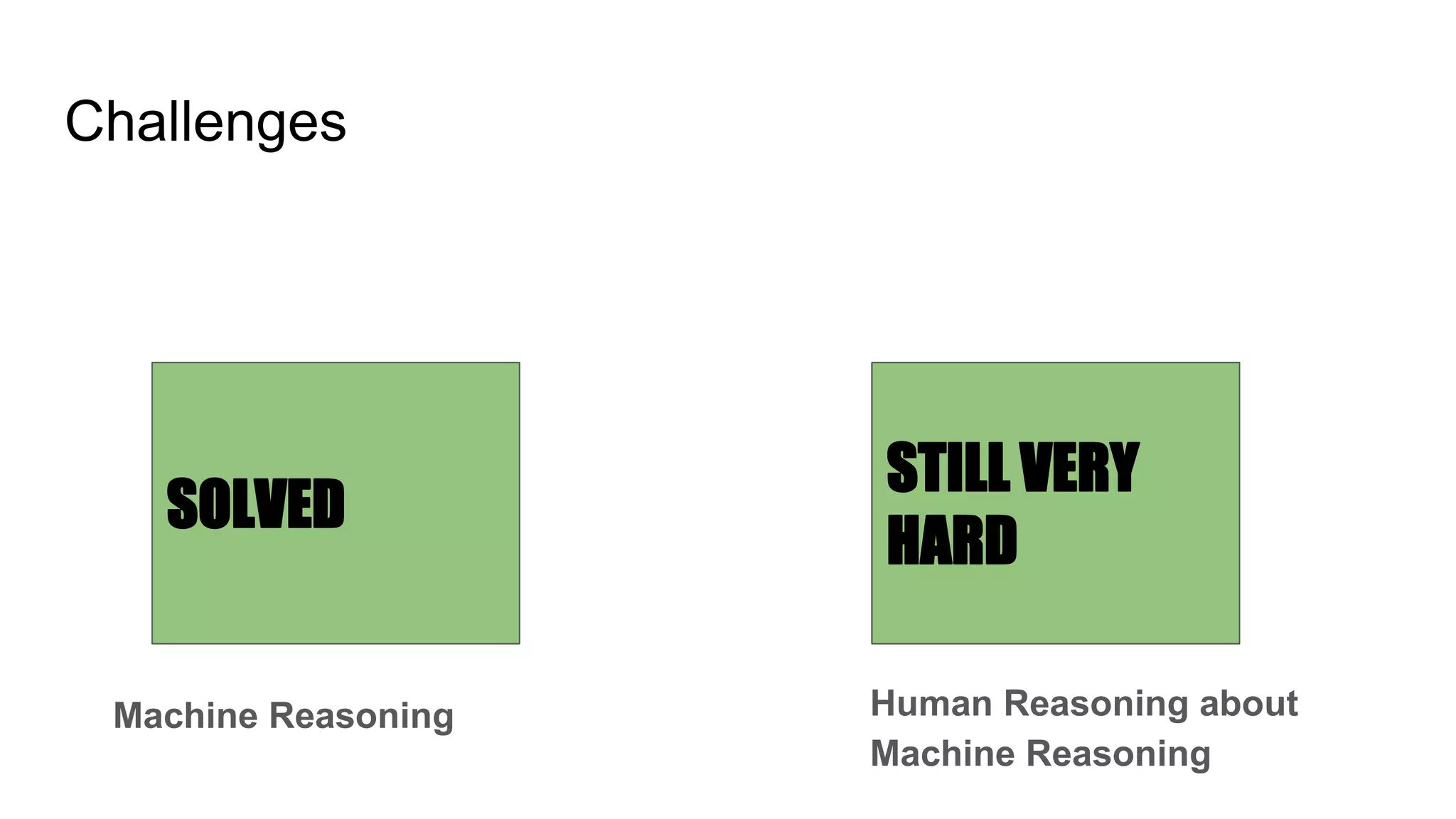
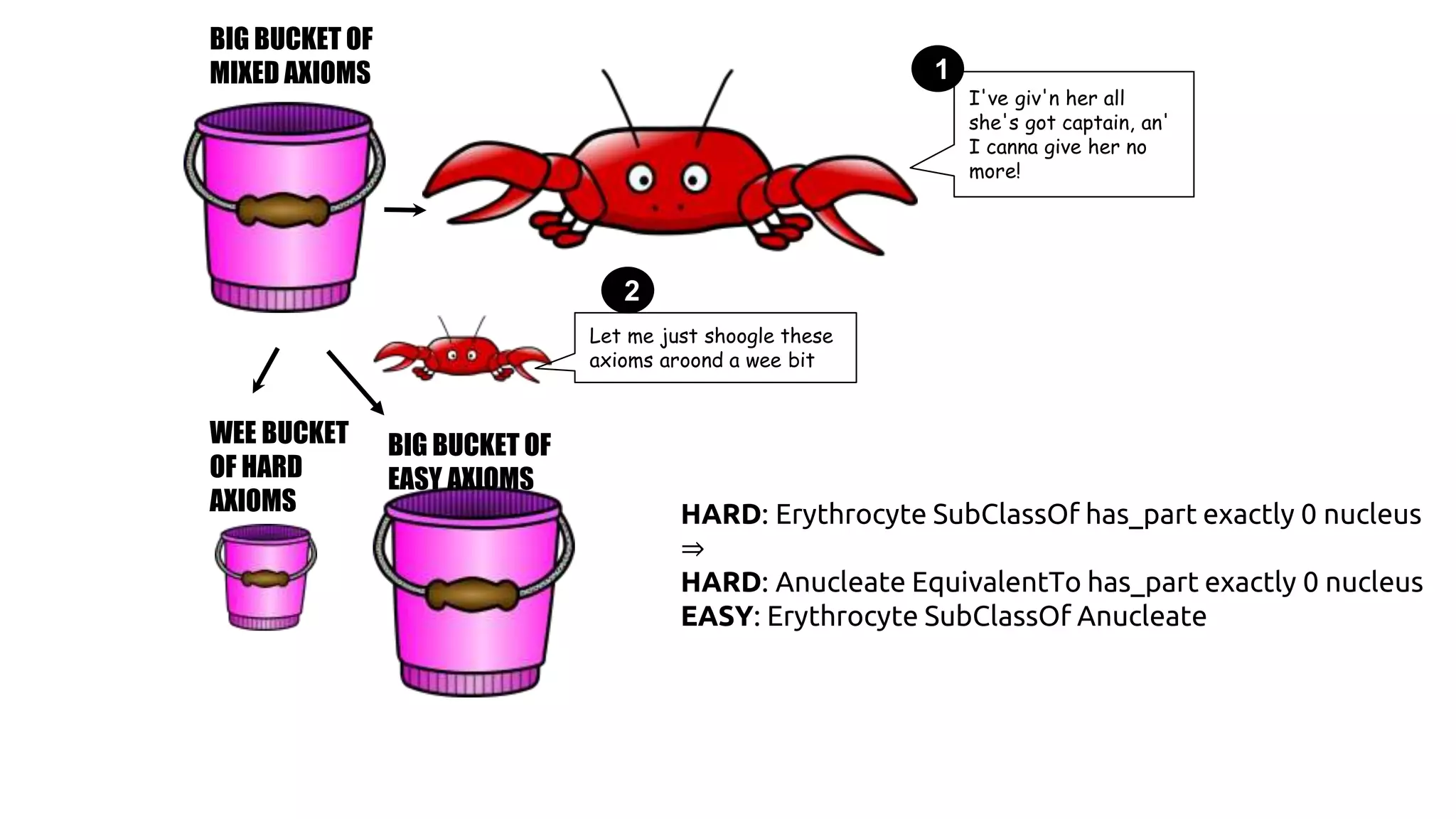
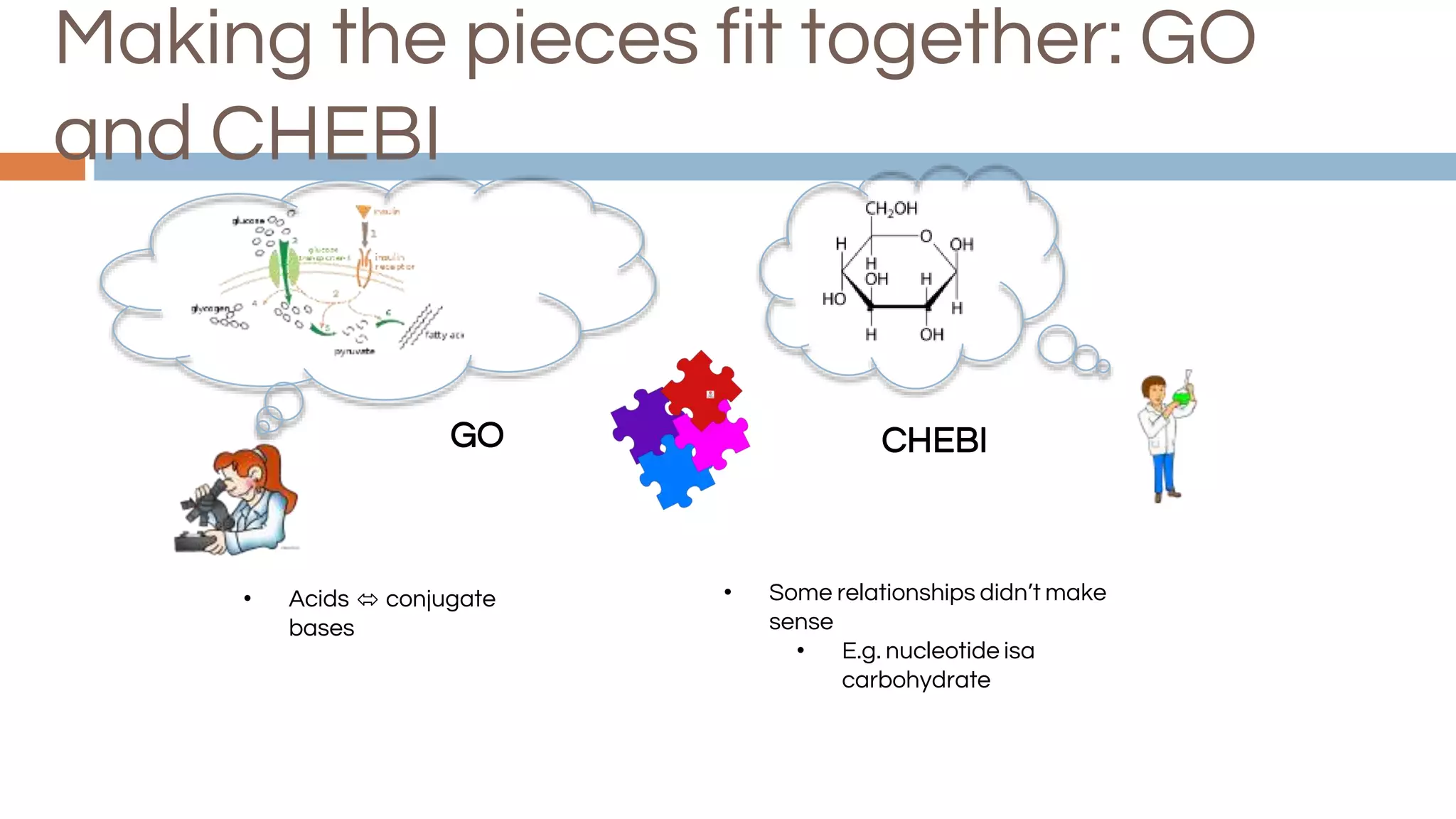
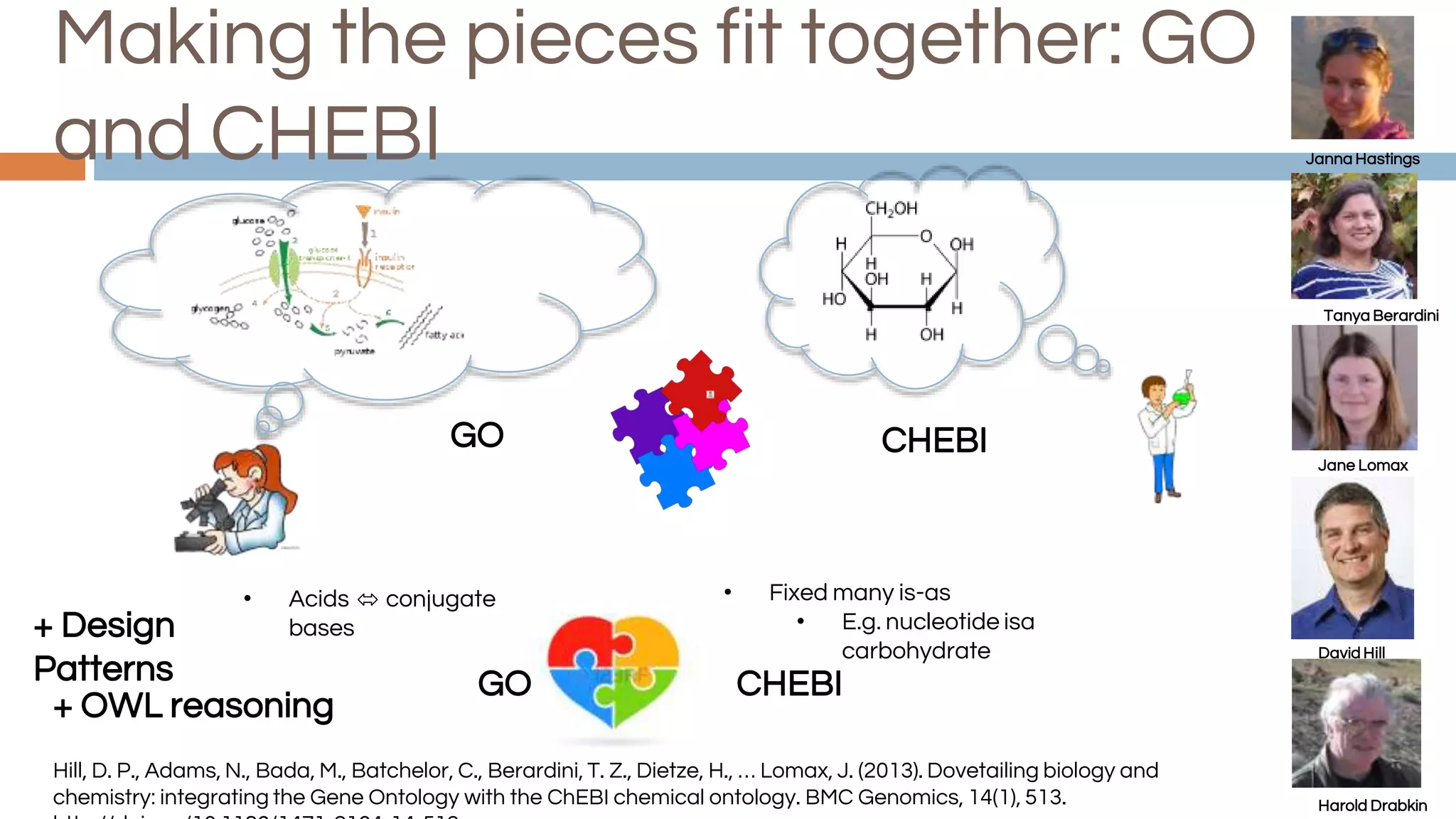
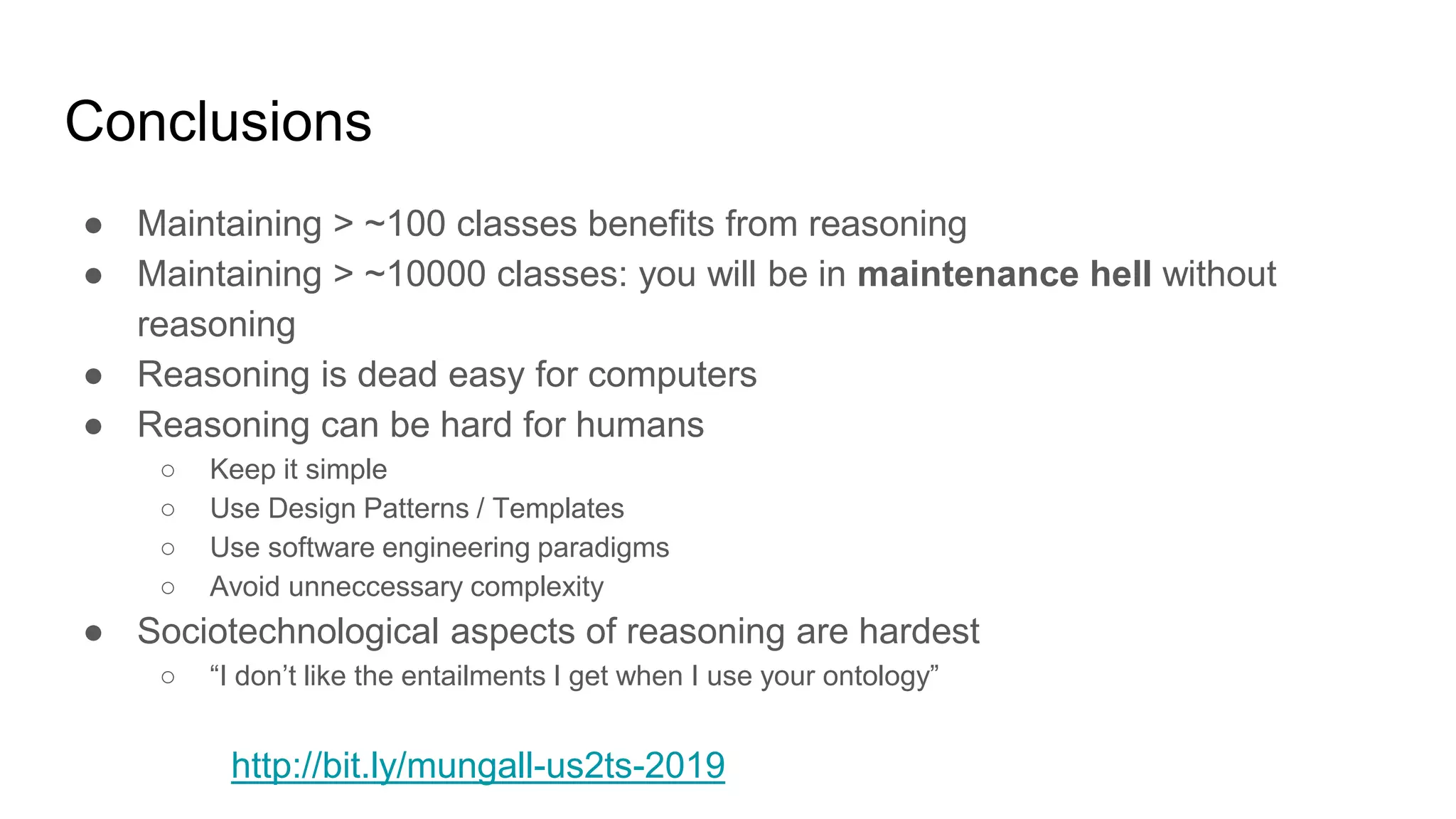
The document discusses the complexities of biological data management, emphasizing the need for modular and well-integrated ontologies to better categorize vast amounts of biological information. It highlights the use of tools and methodologies, such as reasoning and design patterns, to address common annotation errors and to facilitate collaboration in ontology development. Ultimately, it stresses that while machine reasoning is straightforward, the sociotechnological aspects and human reasoning about these ontologies can present significant challenges.