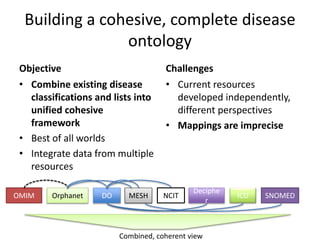
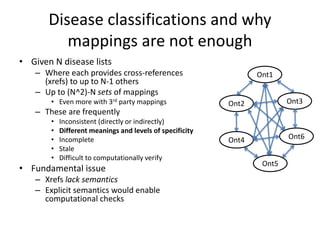
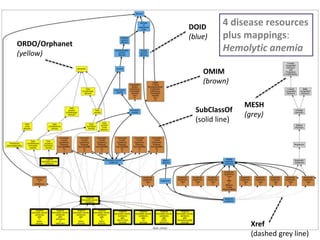
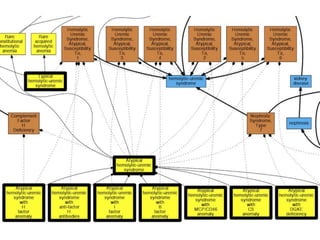
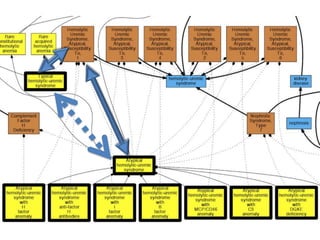
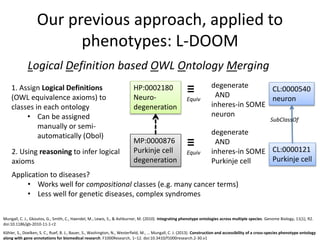
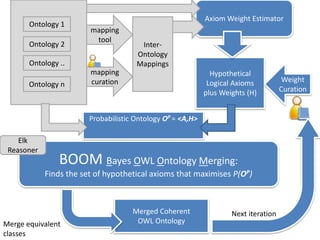
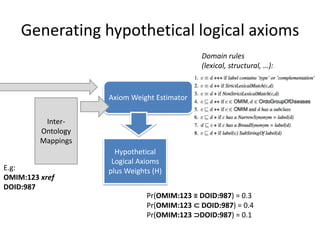
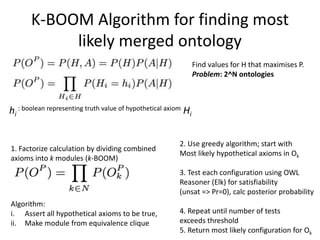
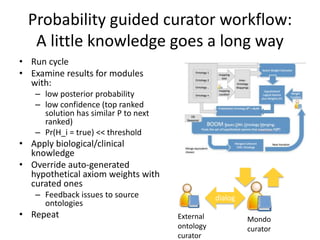
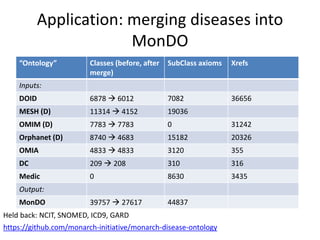
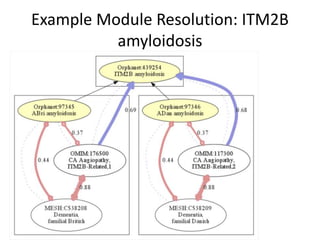
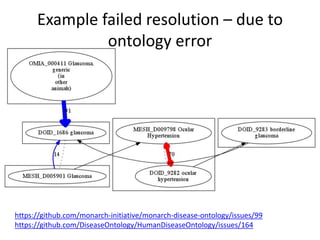
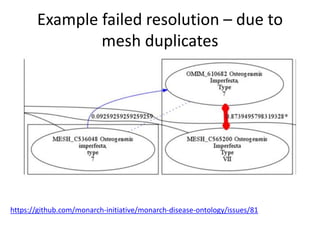
Chris Mungall presented a Bayesian approach called k-BOOM (Bayesian OWL Ontology Merging) to combine existing disease ontologies and lists into a unified framework. k-BOOM generates hypothetical logical mappings between ontologies, estimates weights for each mapping, and uses a greedy algorithm to find the set of mappings that maximizes the probability of the merged ontology. It has been applied to merge several disease ontologies into MonDO (Monarch Disease Ontology). Evaluation found high agreement with held-out data and detection of errors in source ontologies. Next steps include improving weight estimation and evaluation.