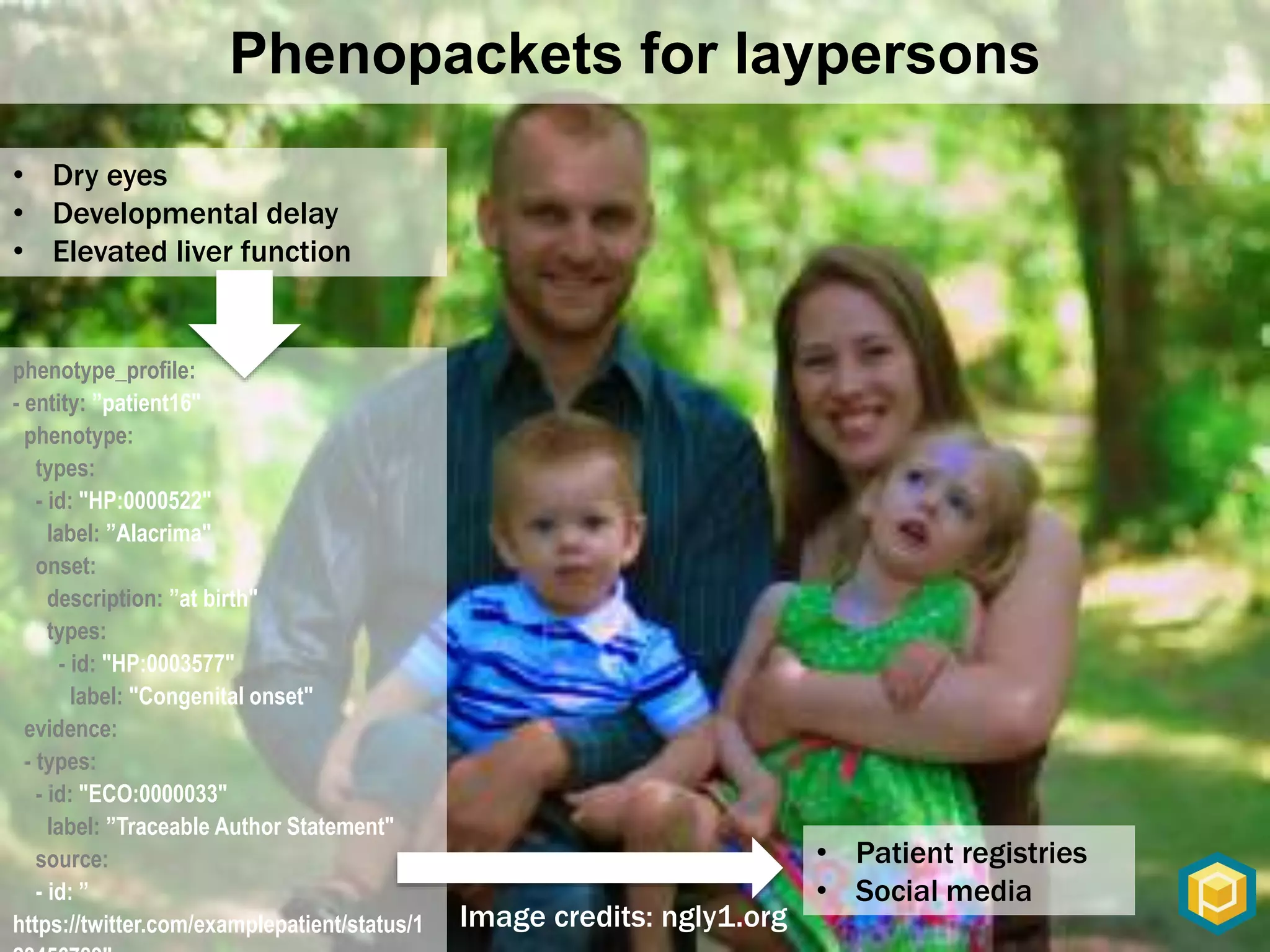
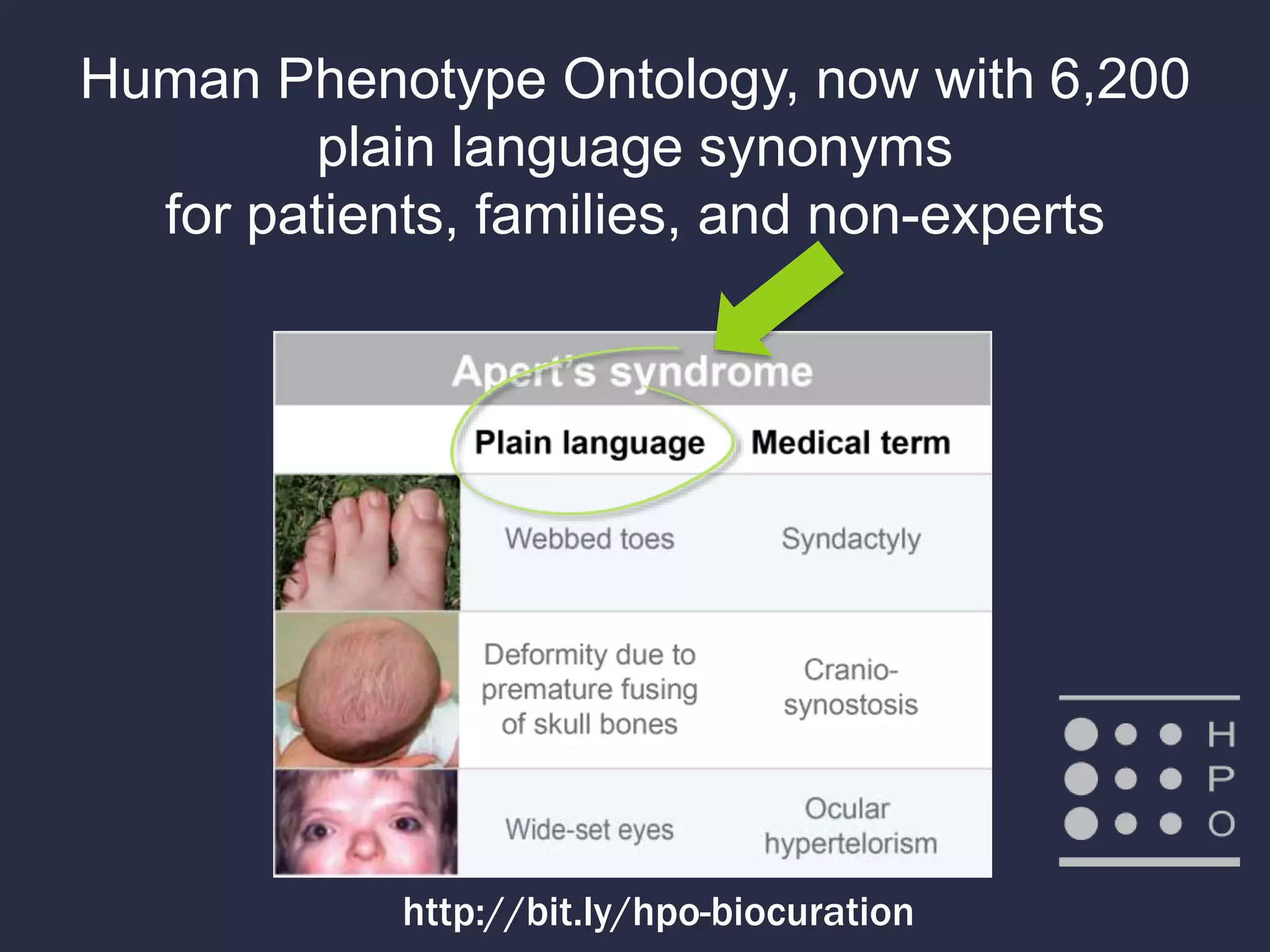
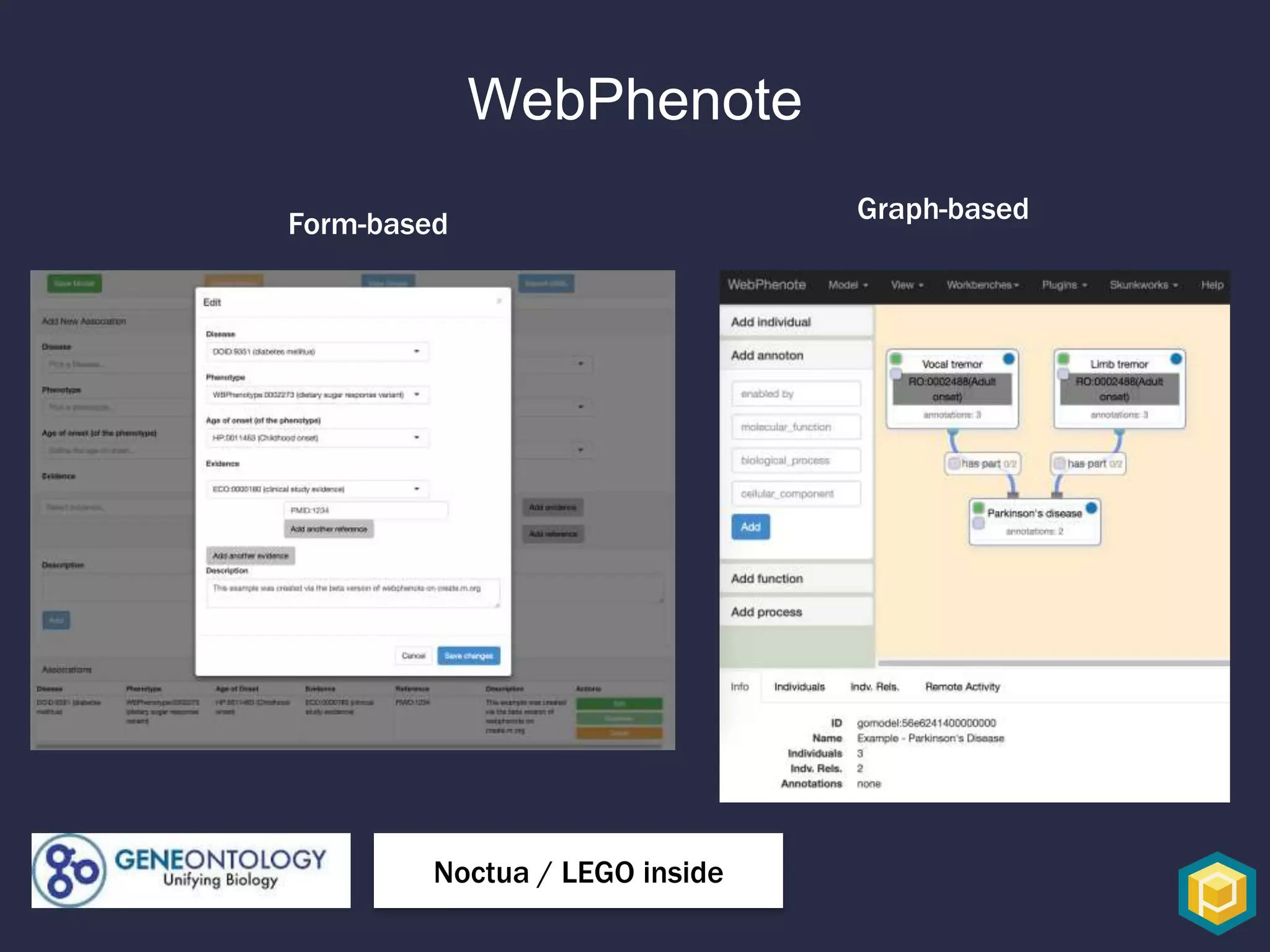
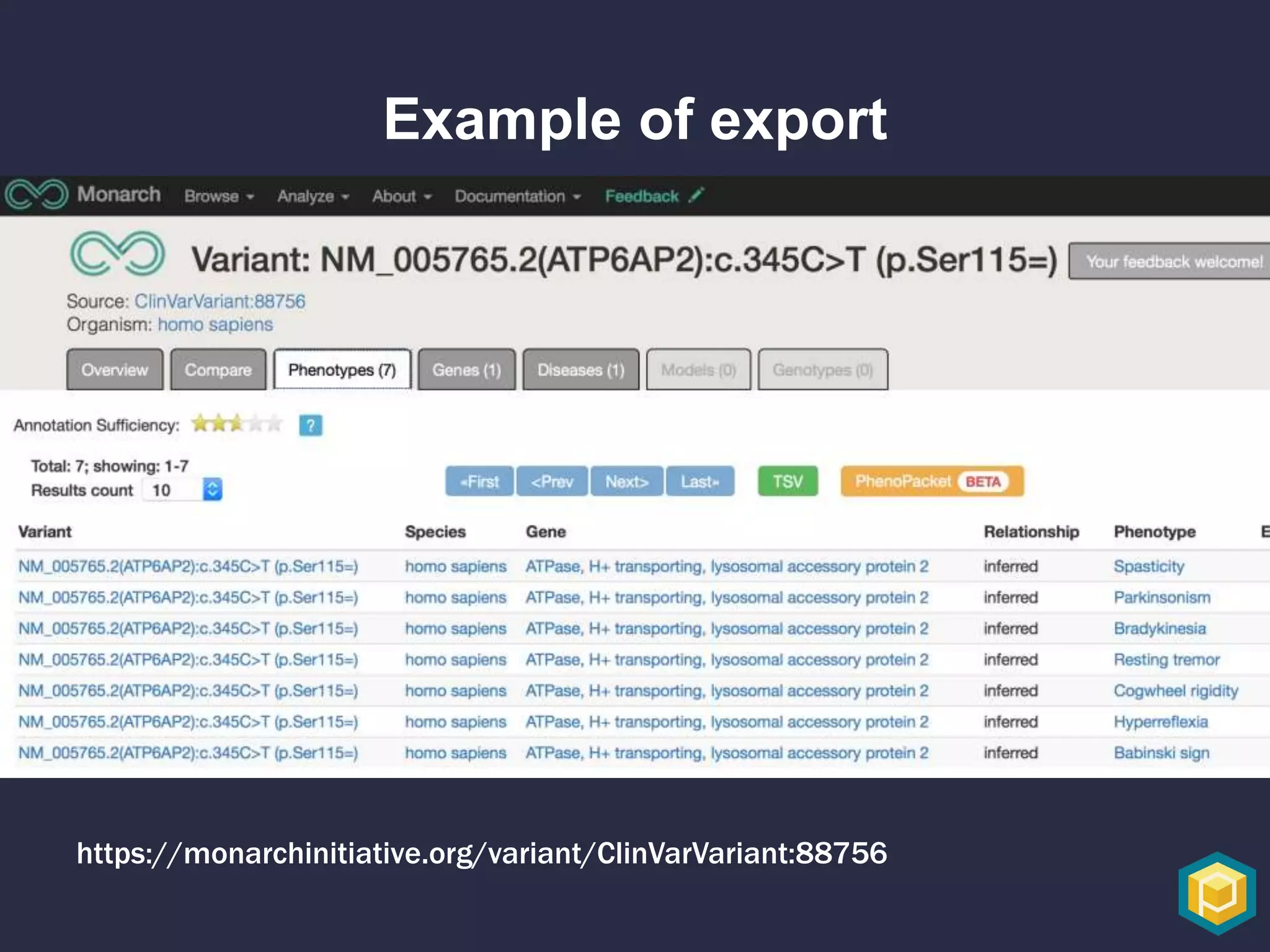
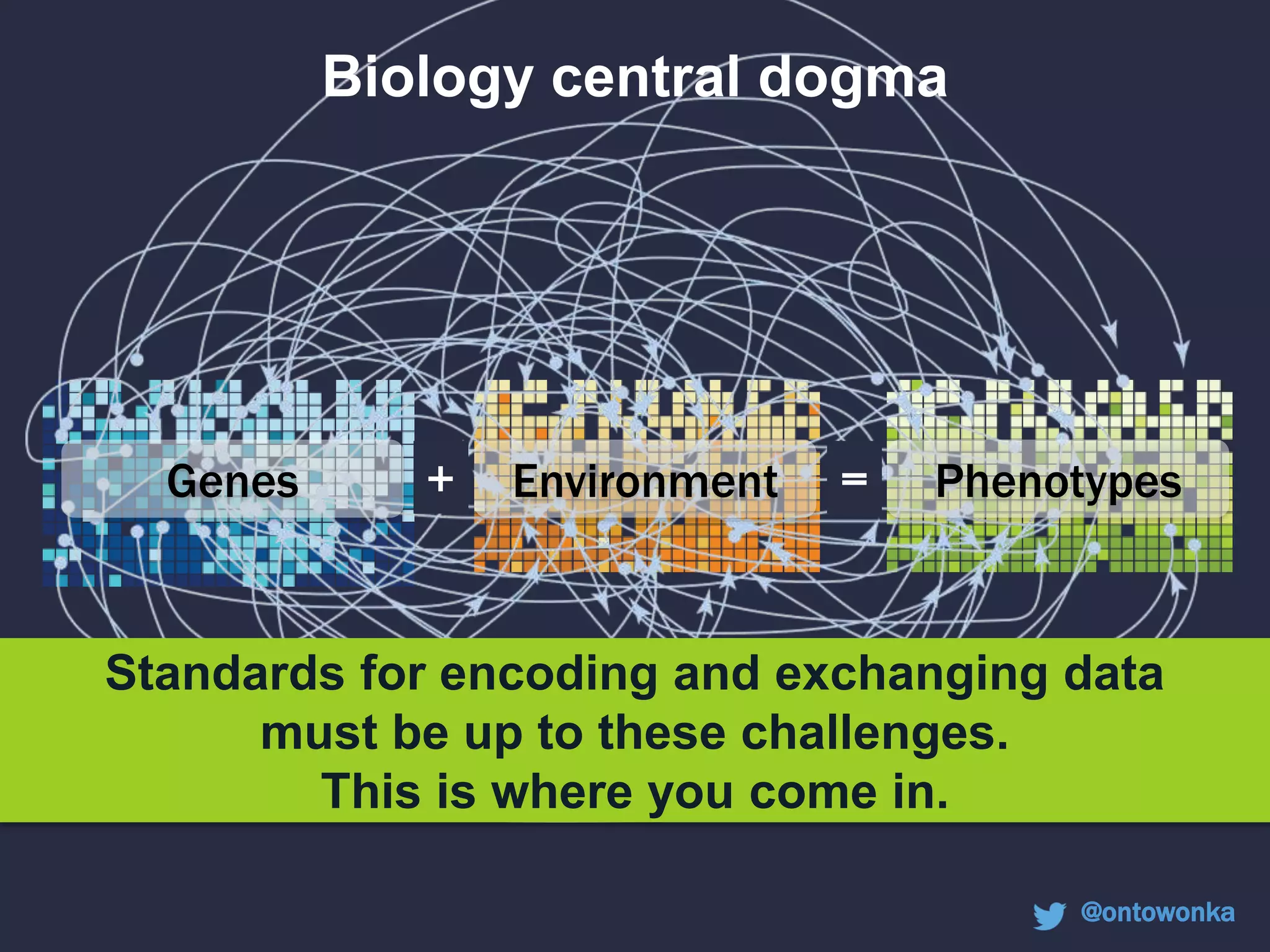
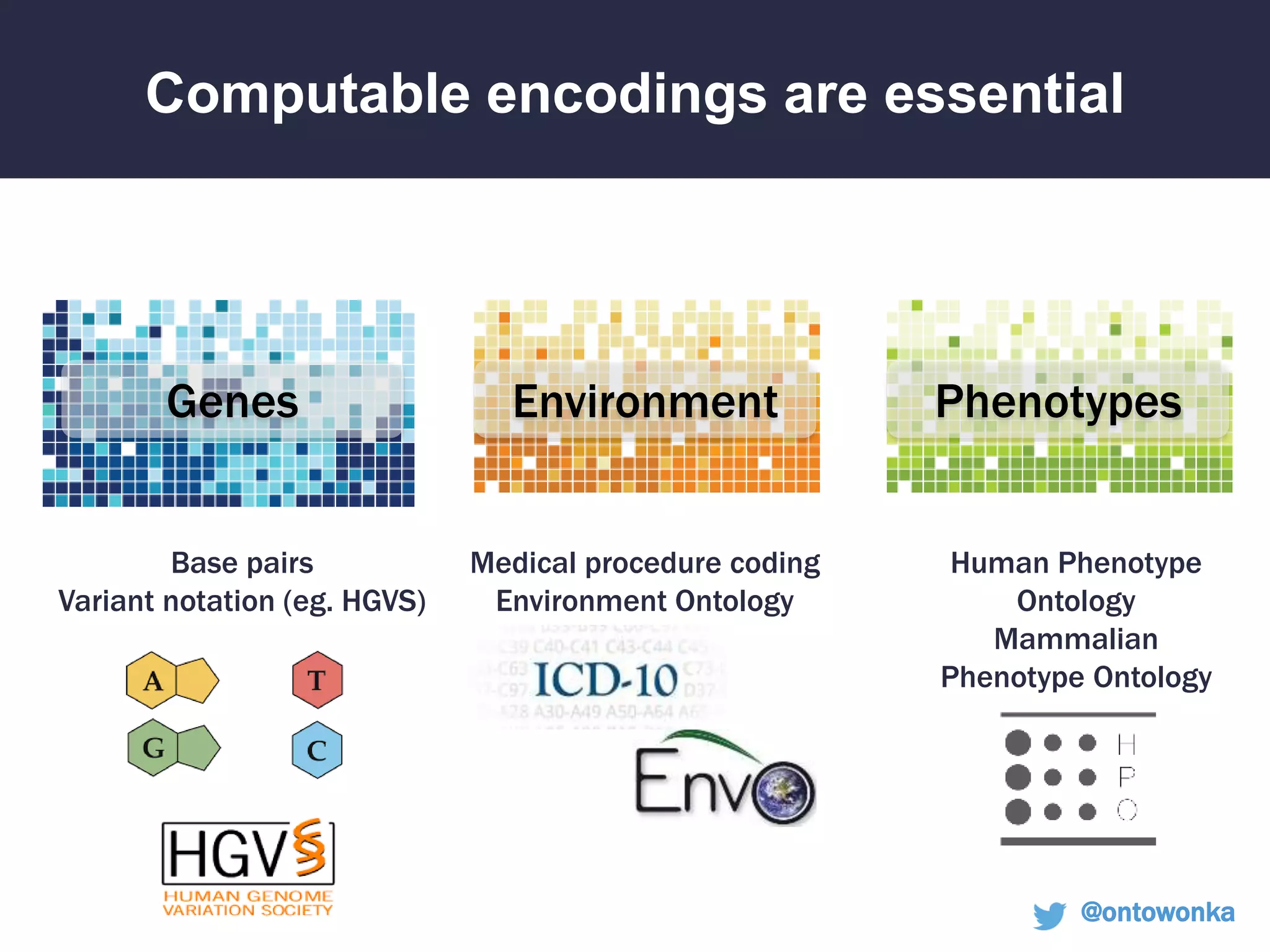
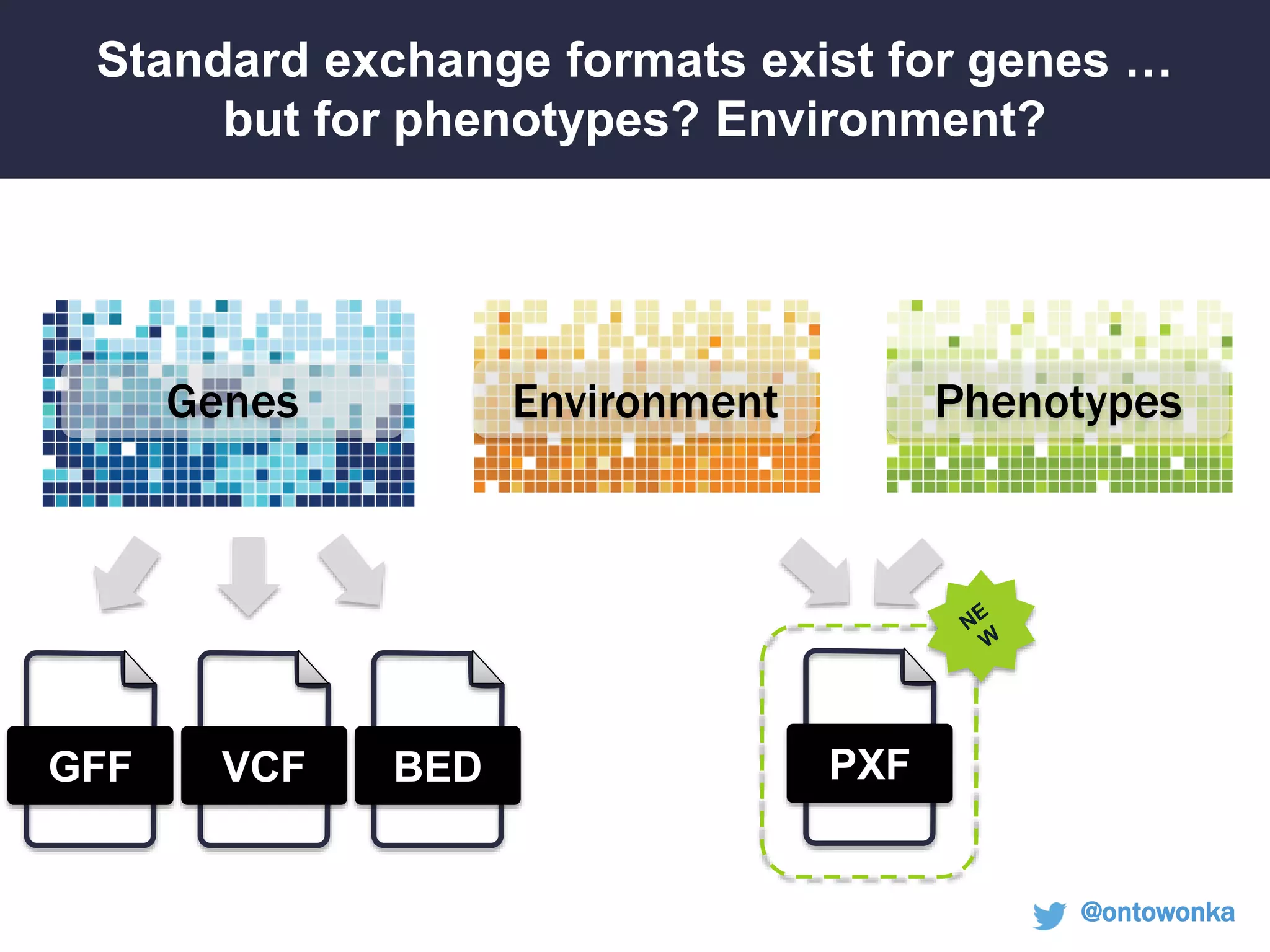
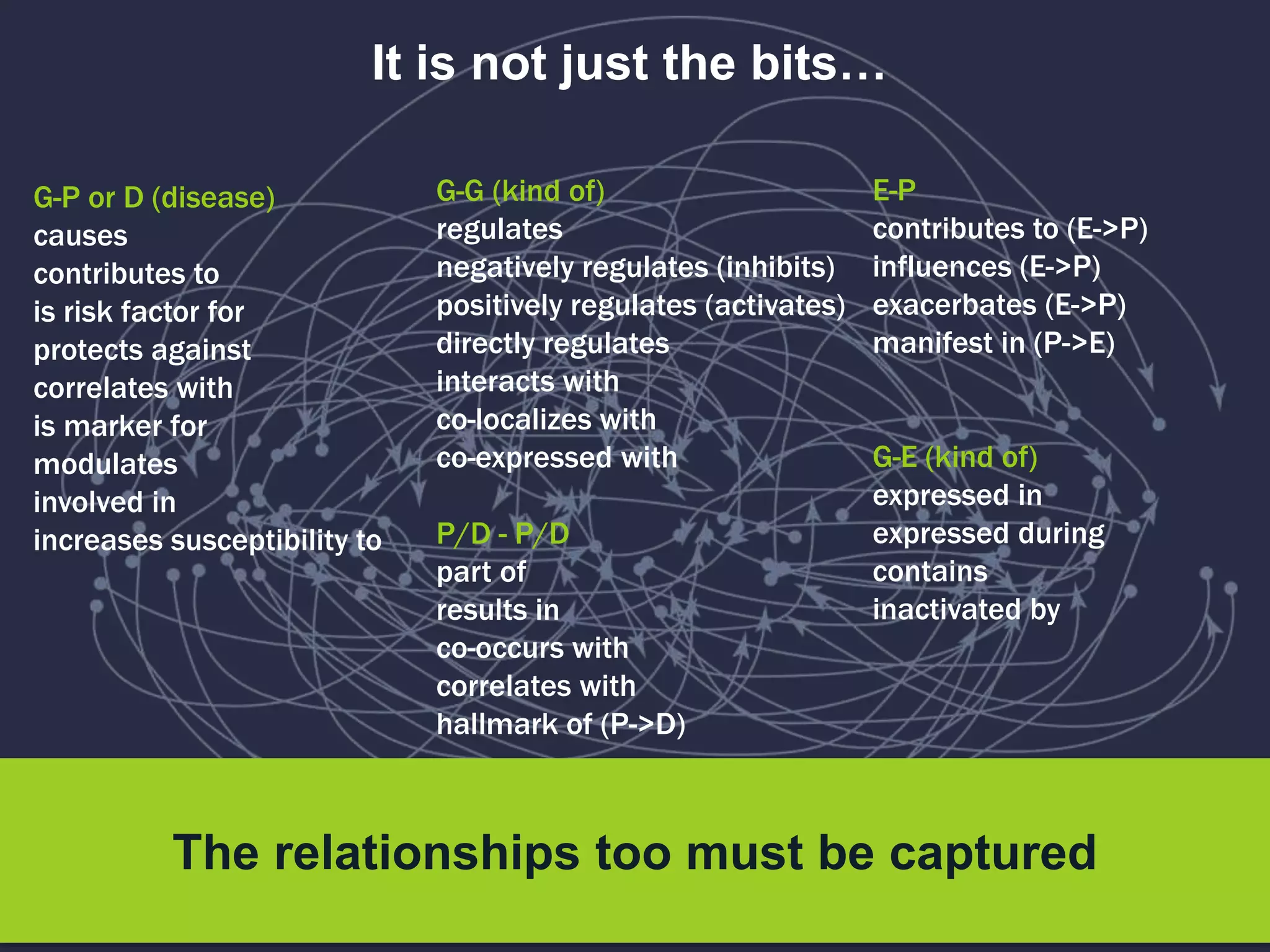
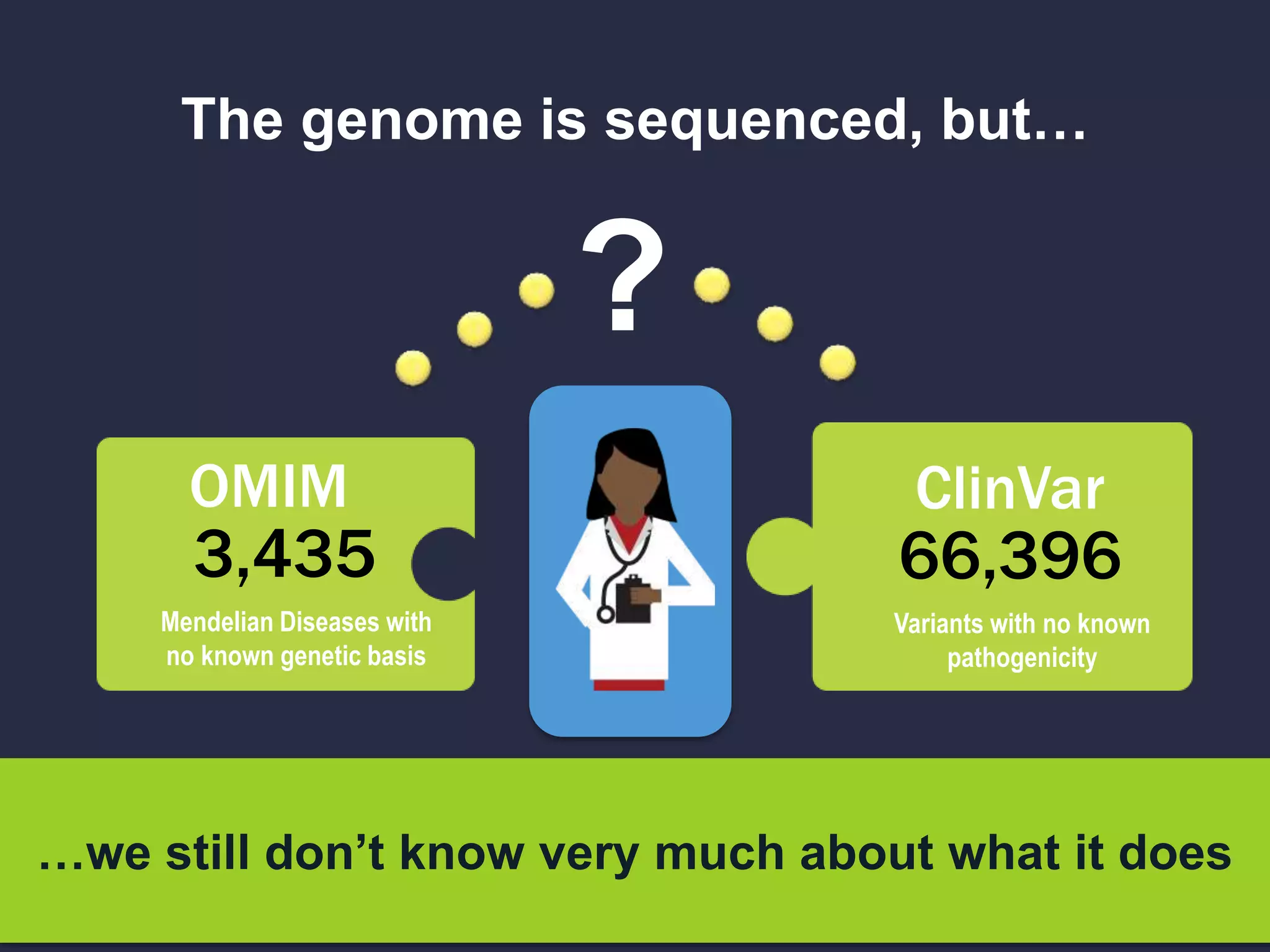
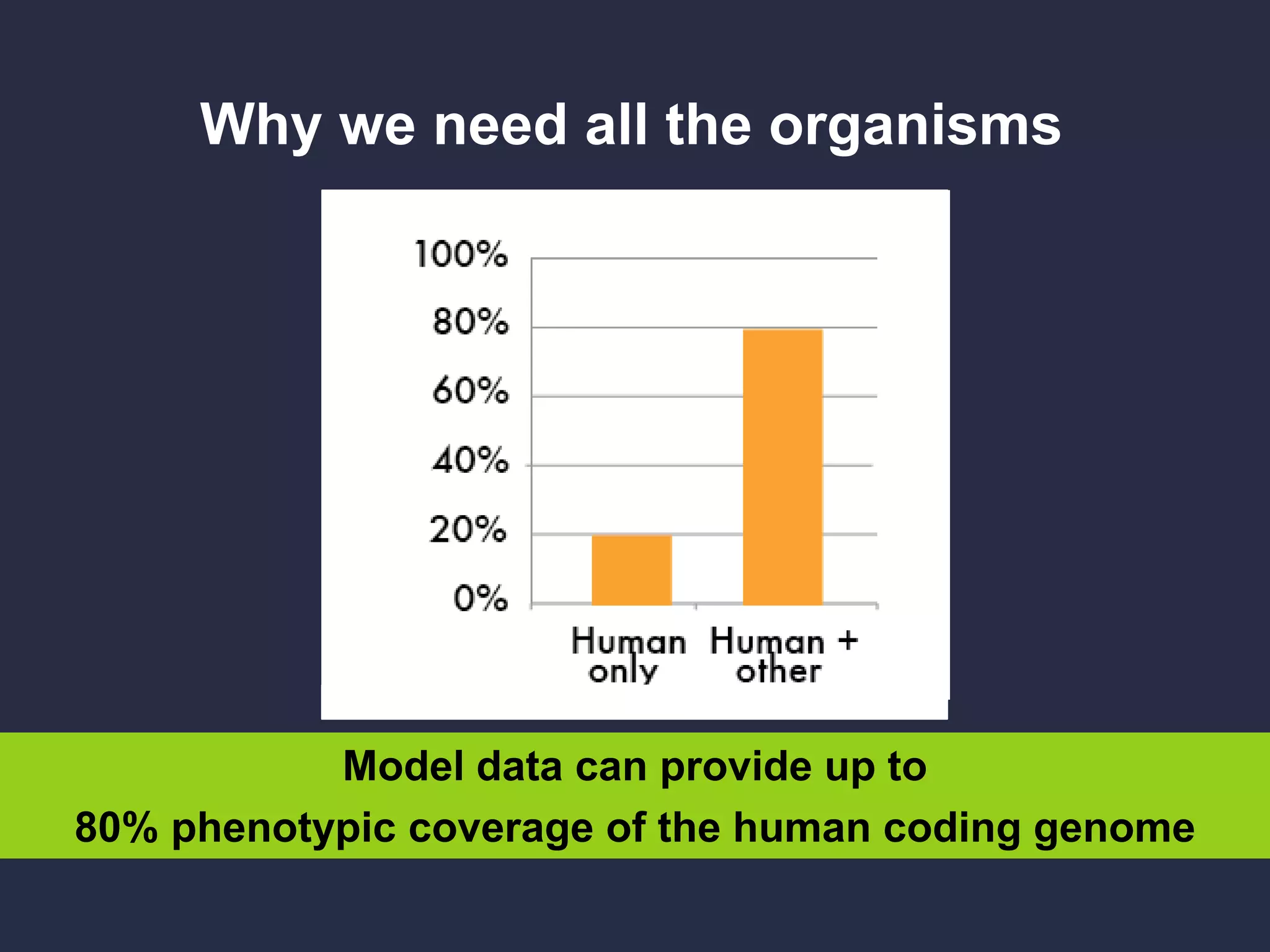
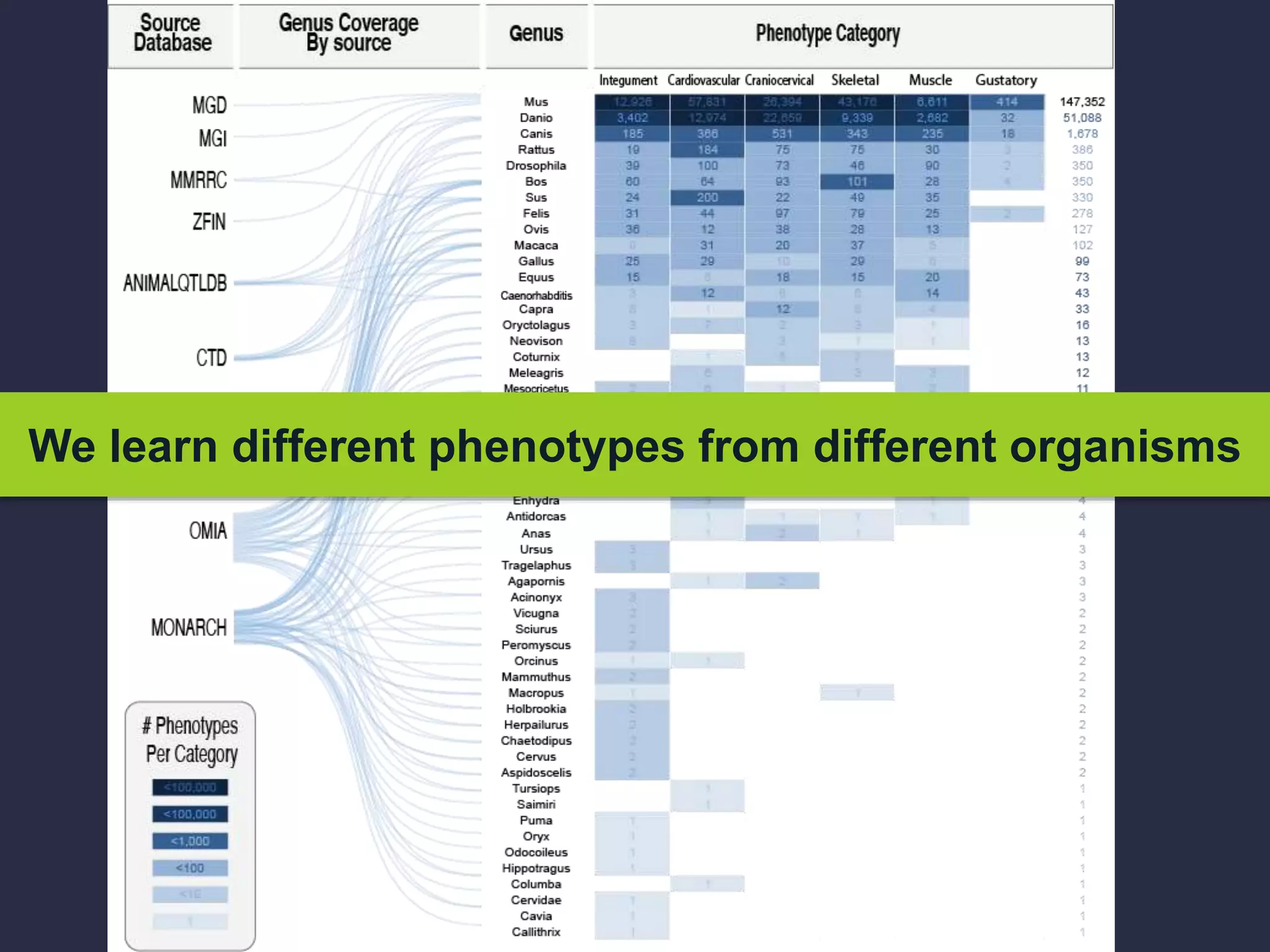
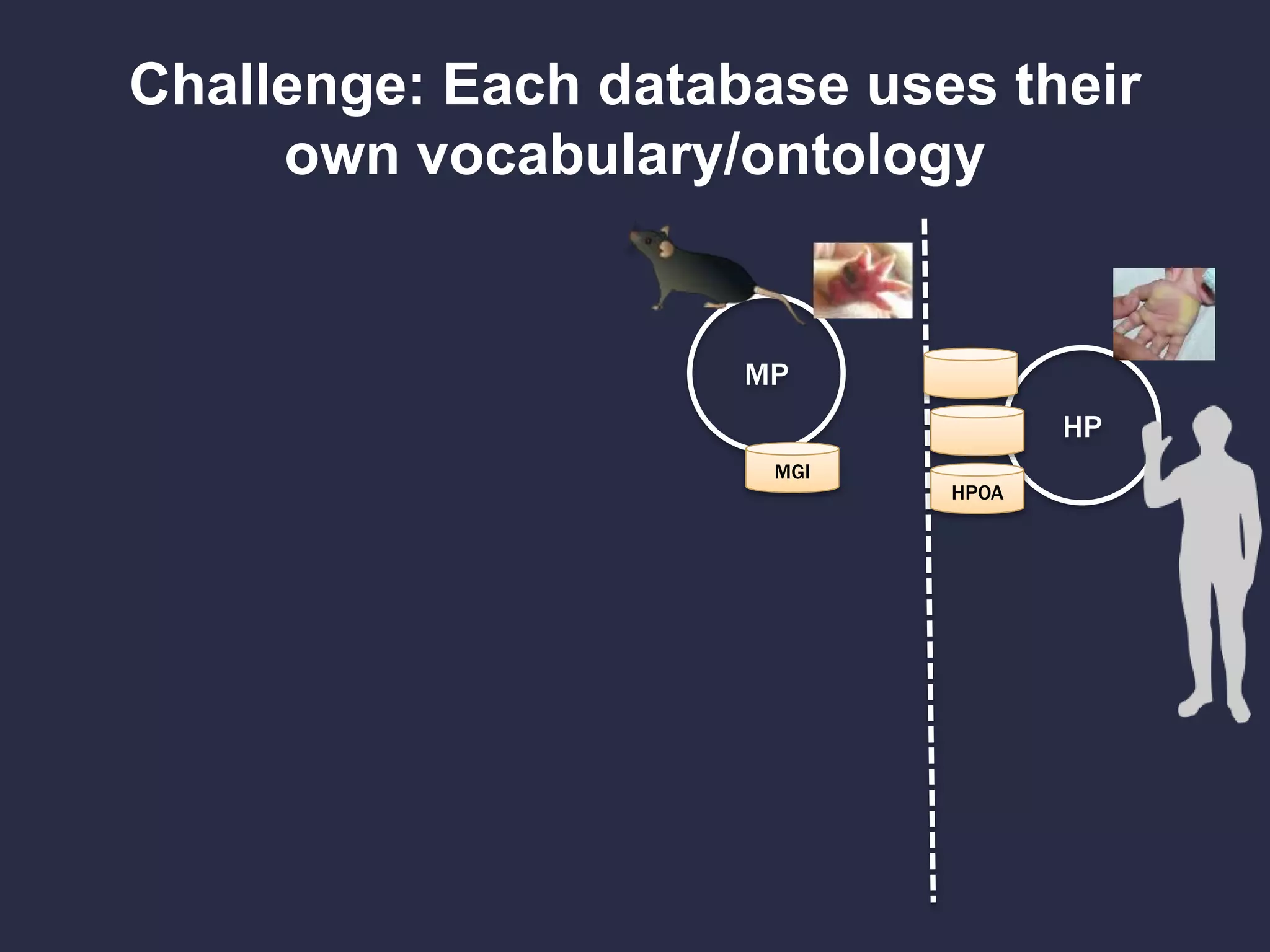
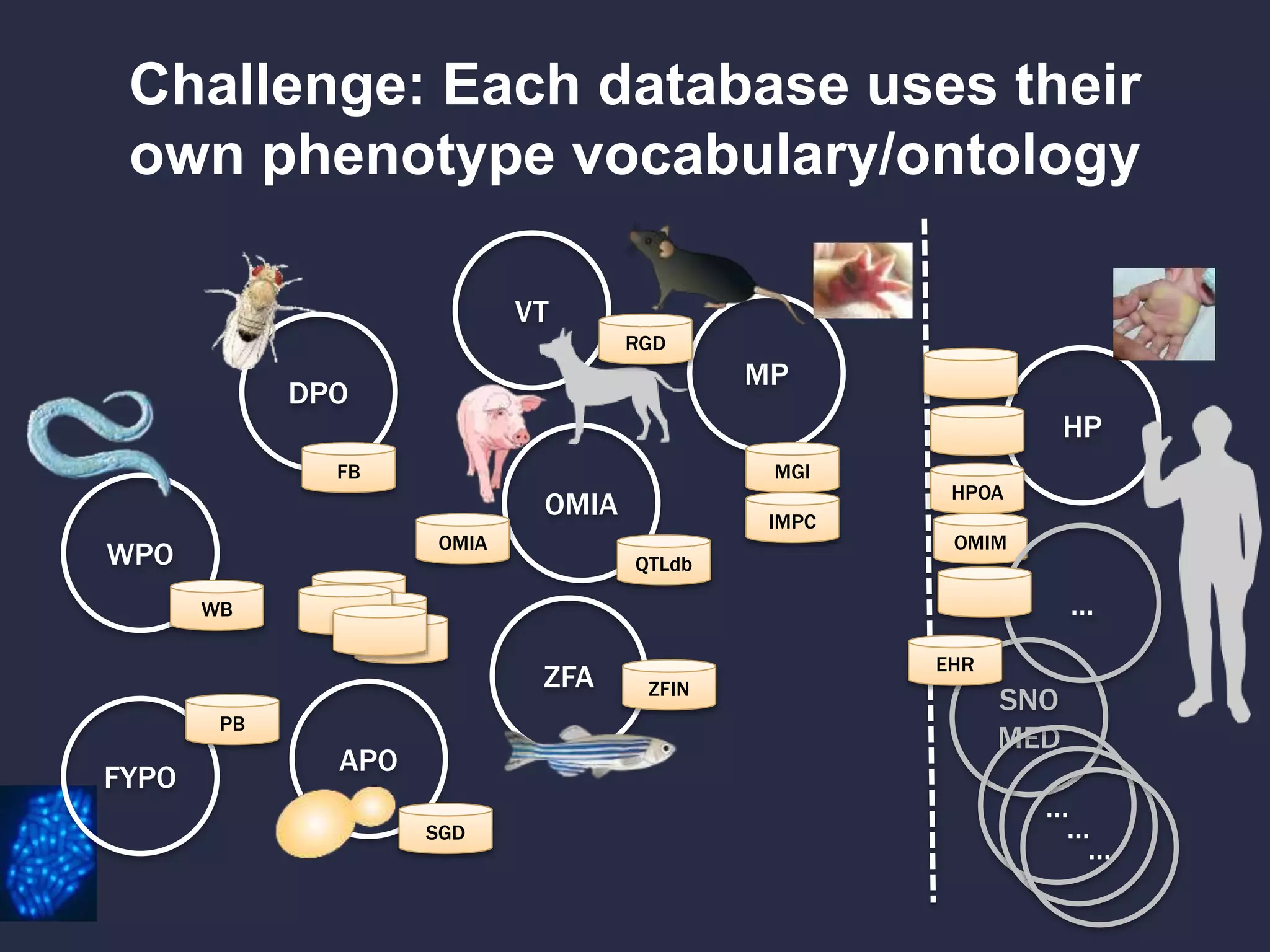
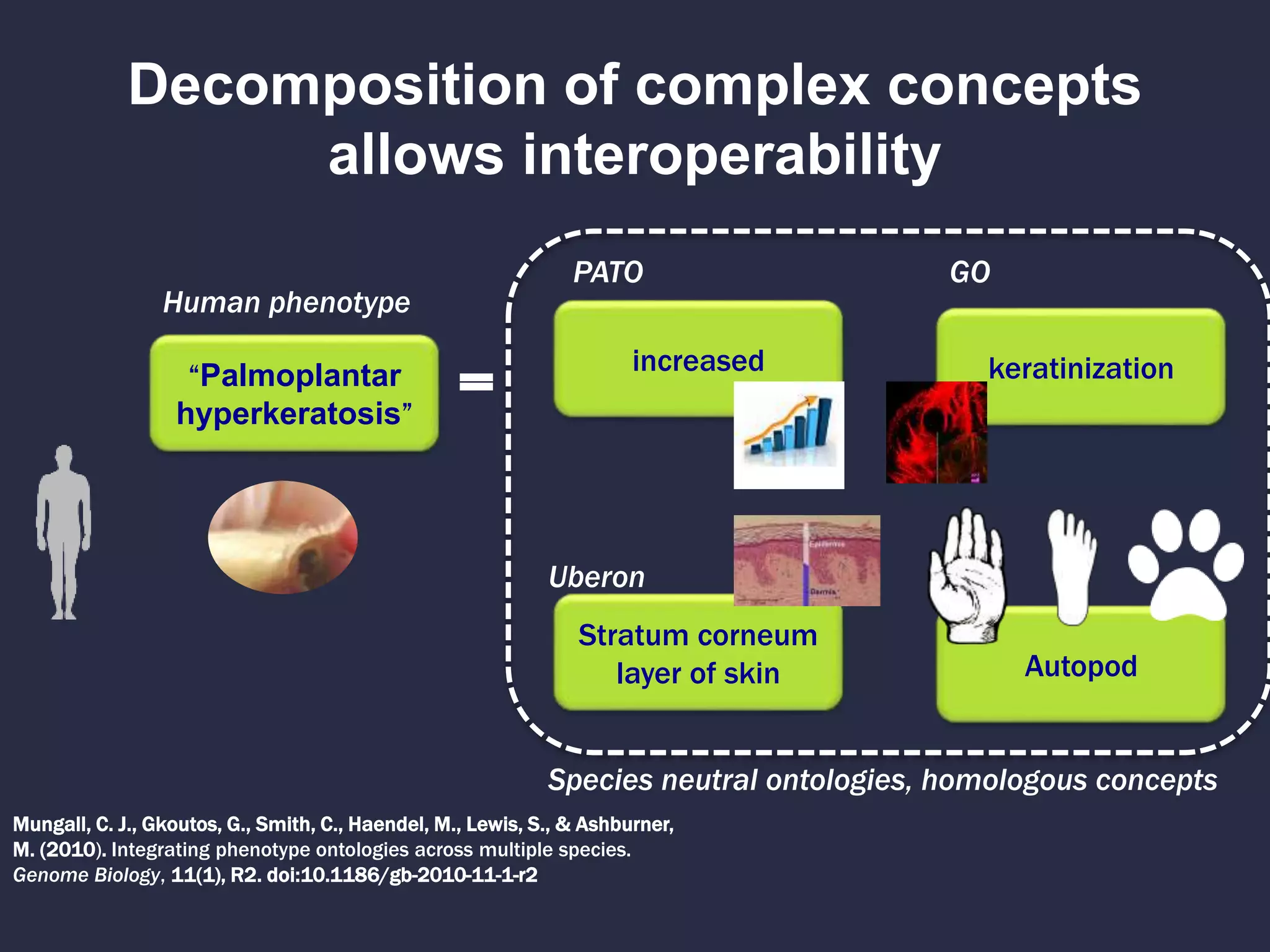
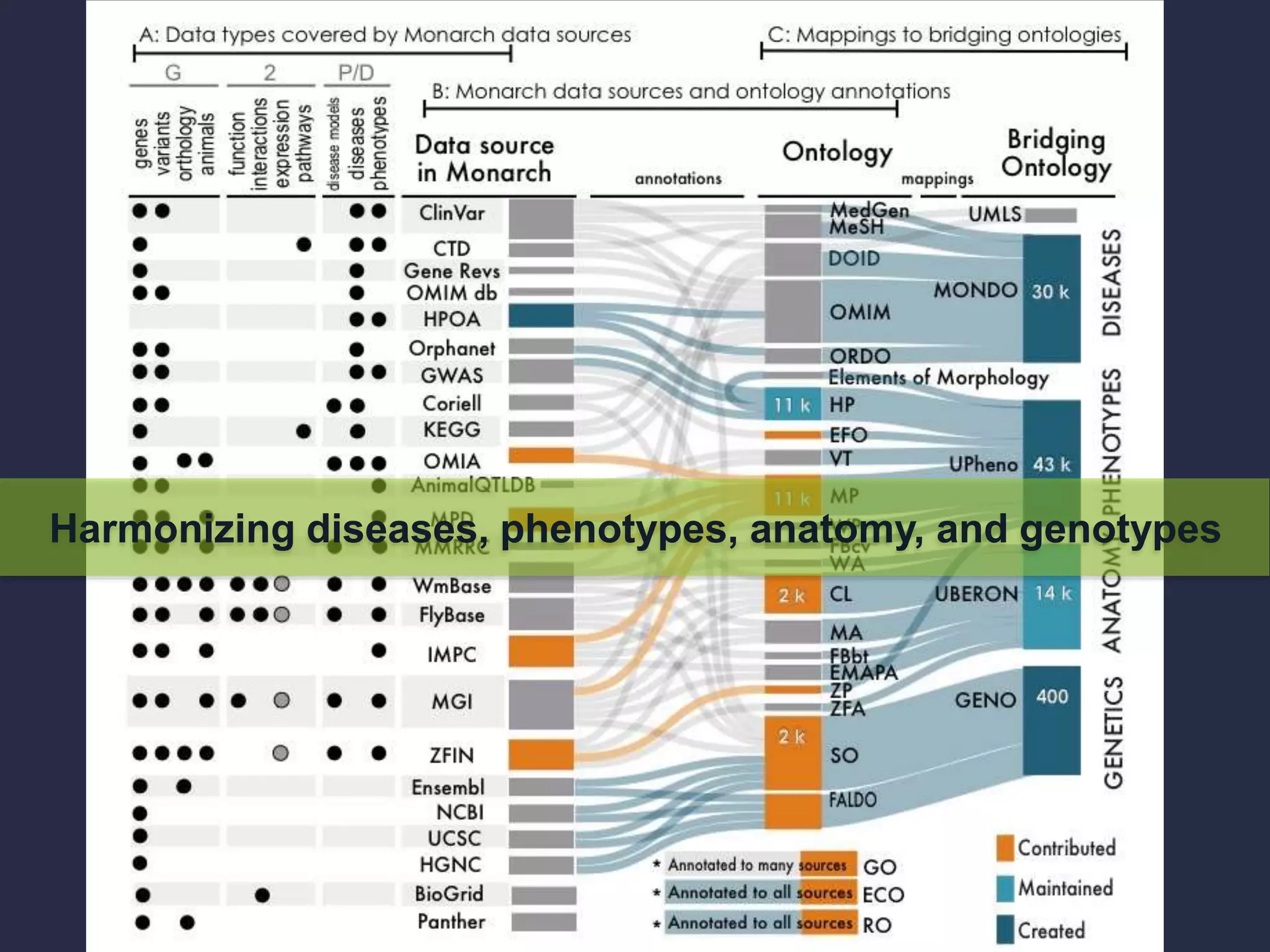
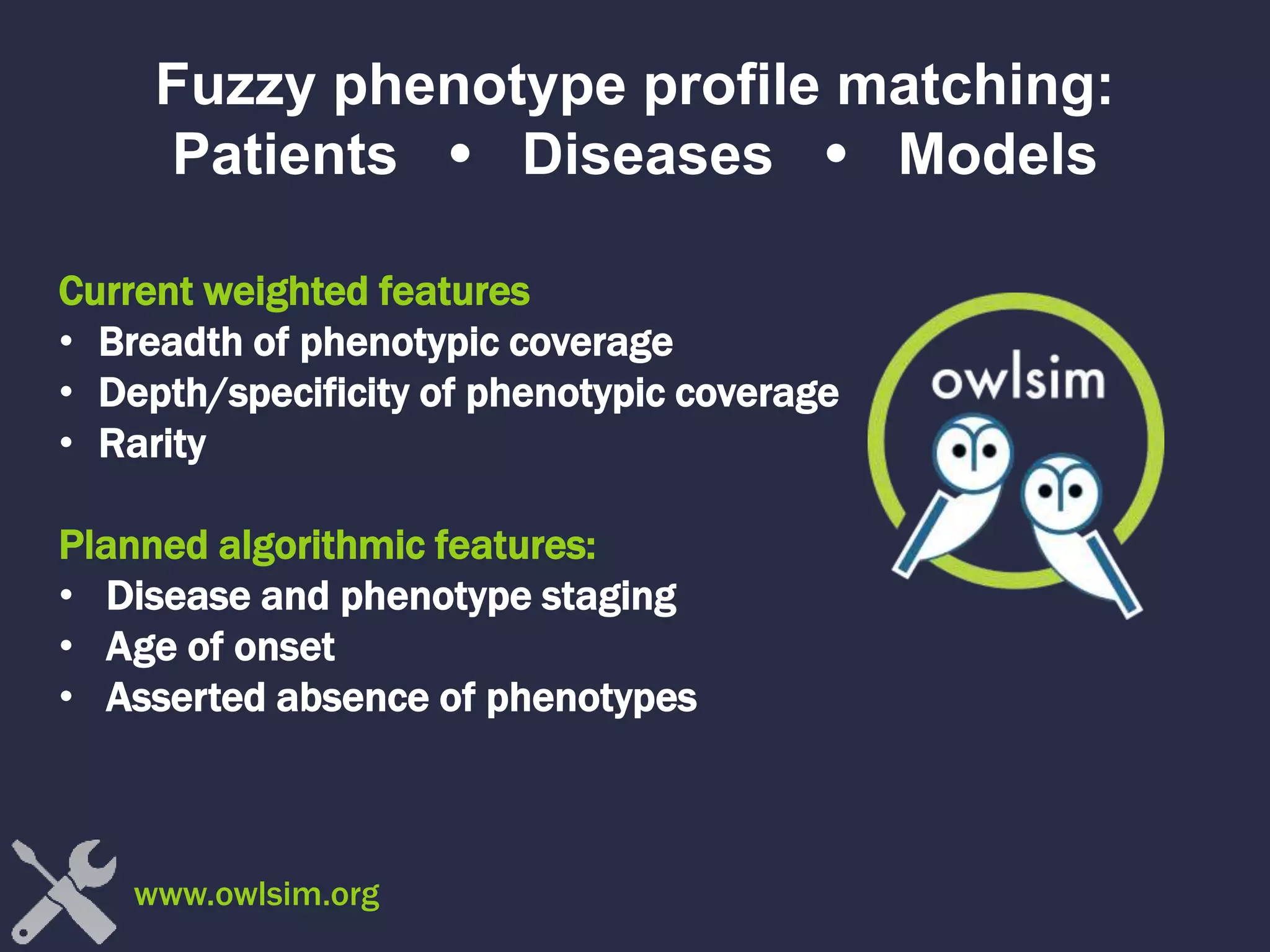
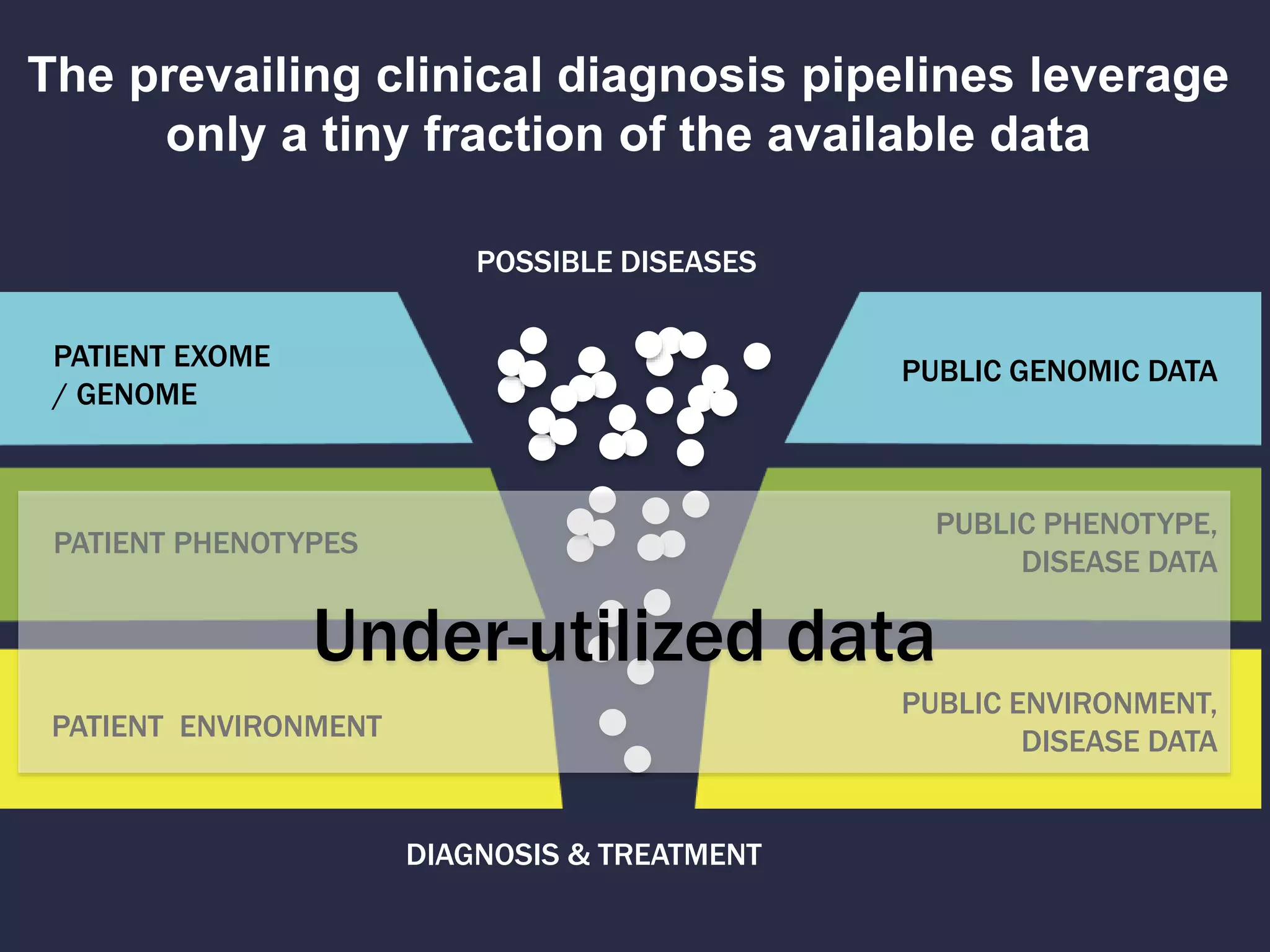
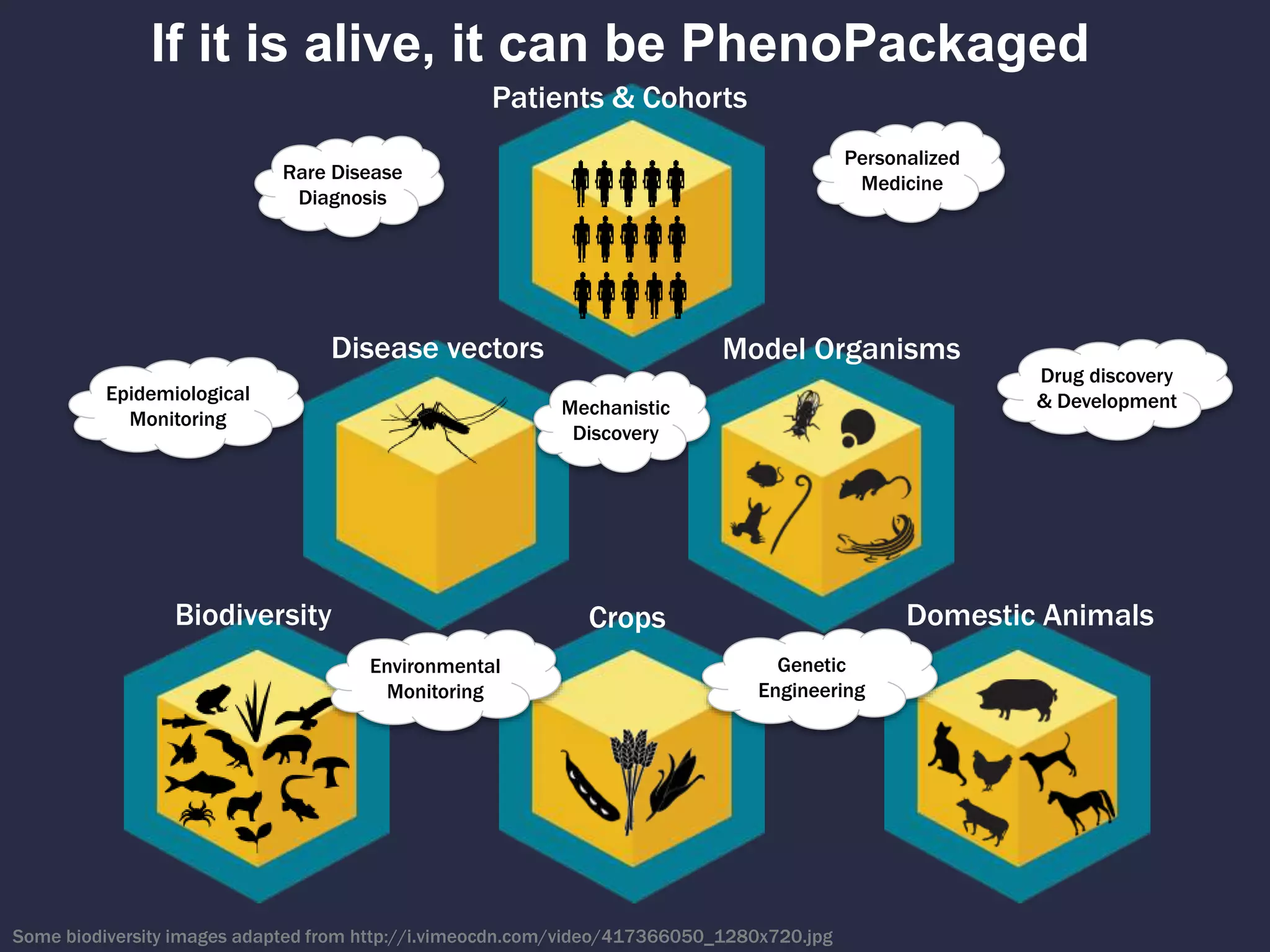
The document discusses the role of 'phenopacketeers' in advancing the understanding and application of phenotype data in fields like medicine and biology. It emphasizes the necessity for standardized encoding and exchange of phenotypic information across various databases to enhance interoperability. The introduction of phenopackets is presented as a solution to create structured phenotype data that can be used universally, benefiting research, clinical diagnostics, and patient care.


![B6.Cg-Alms1foz/fox/J
increased weight,
adipose tissue volume,
glucose homeostasis altered
ALSM1(NM_015120.4)
[c.10775delC] + [-]
GENOTYPE
PHENOTYPE
obesity,
diabetes mellitus,
insulin resistance
increased food intake,
hyperglycemia,
insulin resistance
kcnj11c14/c14; insrt143/+(AB)
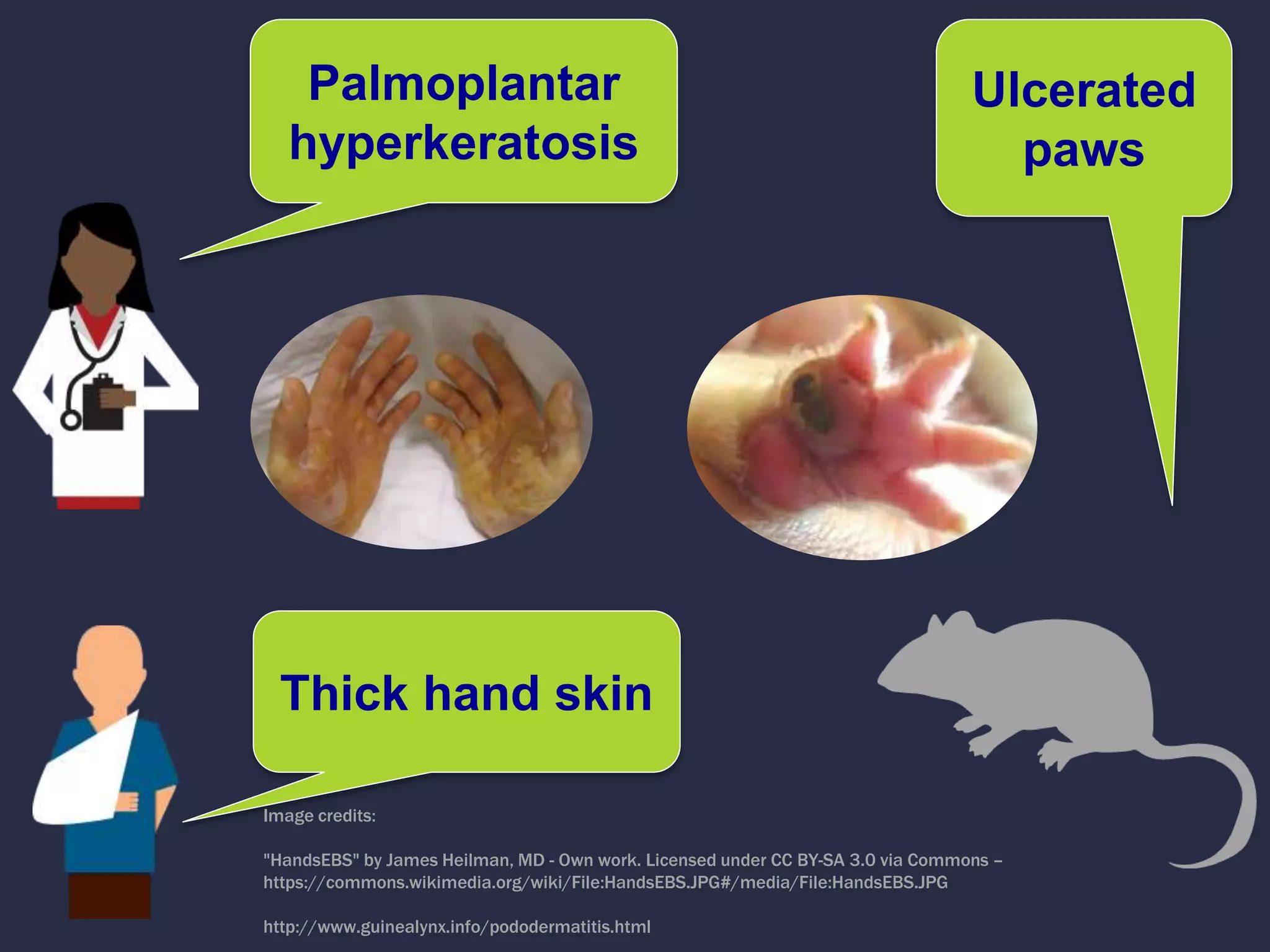
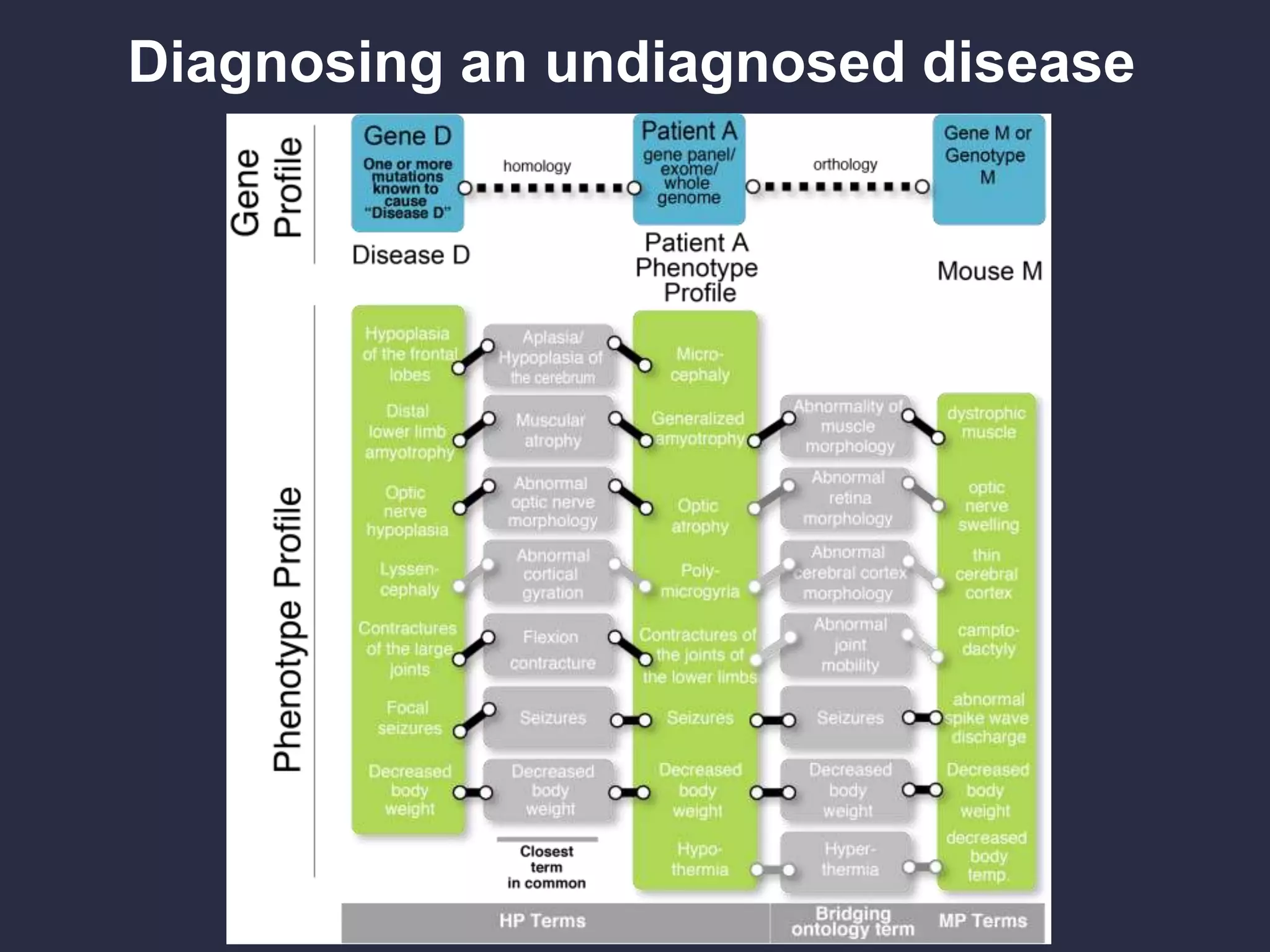
Can we use model phenotypes to
inform genetic mechanisms of disease?
???](https://image.slidesharecdn.com/haendelbiocurationkeynote2016-04-14final-160428005146/75/Why-the-world-needs-phenopacketeers-and-how-to-be-one-11-2048.jpg)





![What is in a PhenoPacket?
This is “Maru”,
a 4-year-old, male
cat of the Scottish
Fold breed
abnormal
sheltering behavior
[MP:0014039]
(onset at birth)
Biography
Phenotypes
&qualifiers
youtube.com/user
/mugumogu
Weighs 6kg
Measurements
Source](https://image.slidesharecdn.com/haendelbiocurationkeynote2016-04-14final-160428005146/75/Why-the-world-needs-phenopacketeers-and-how-to-be-one-27-2048.jpg)