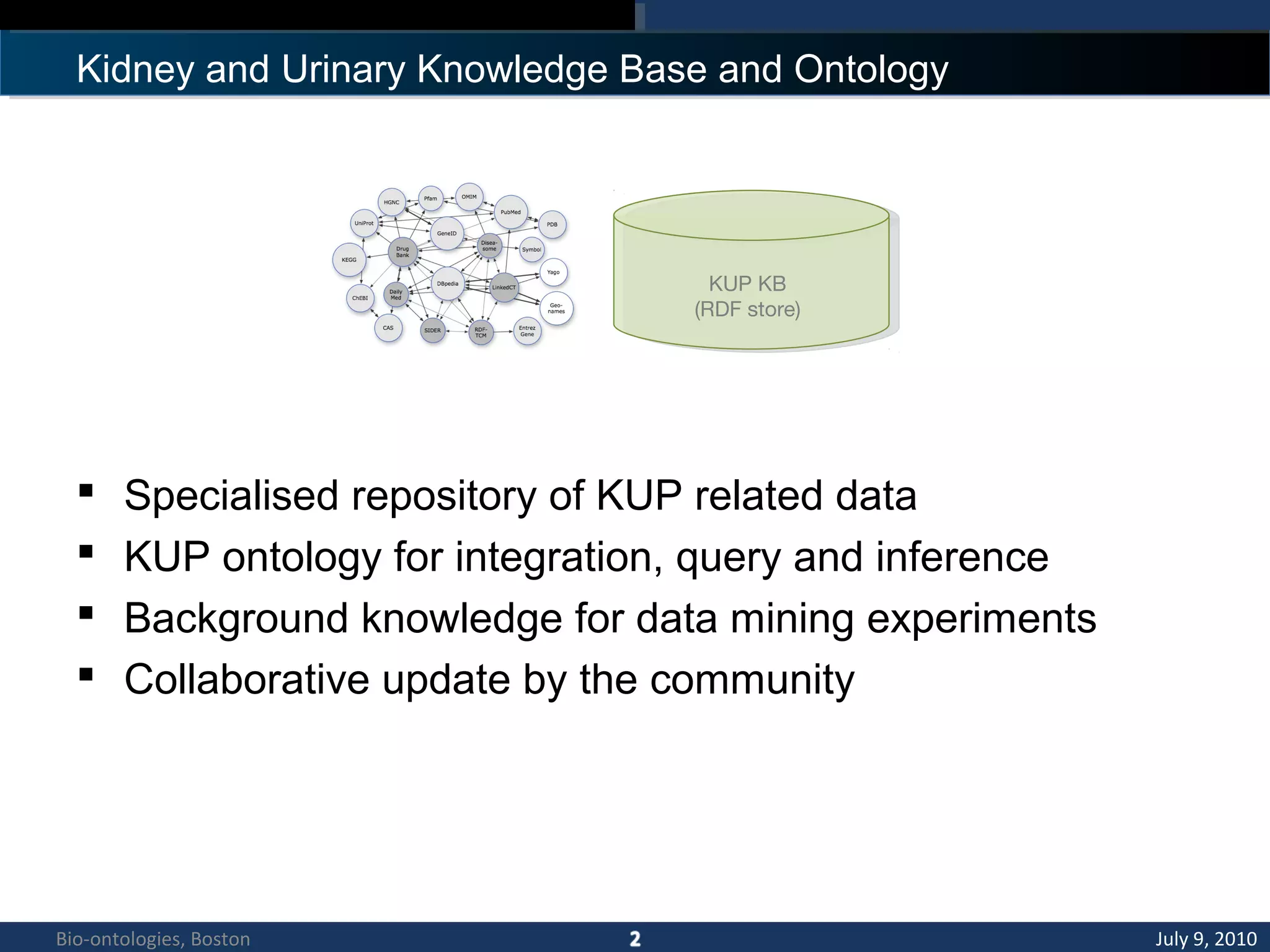
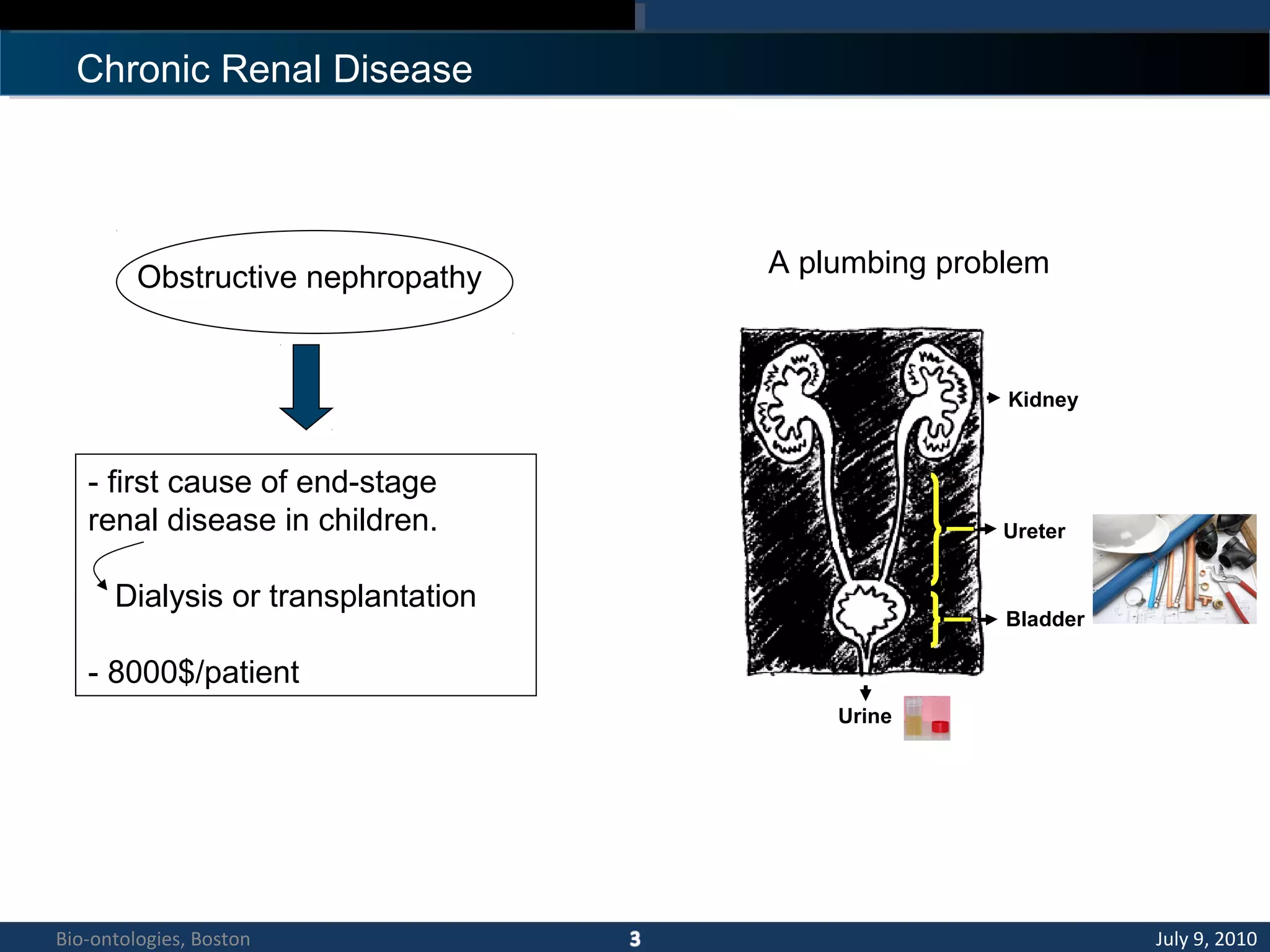
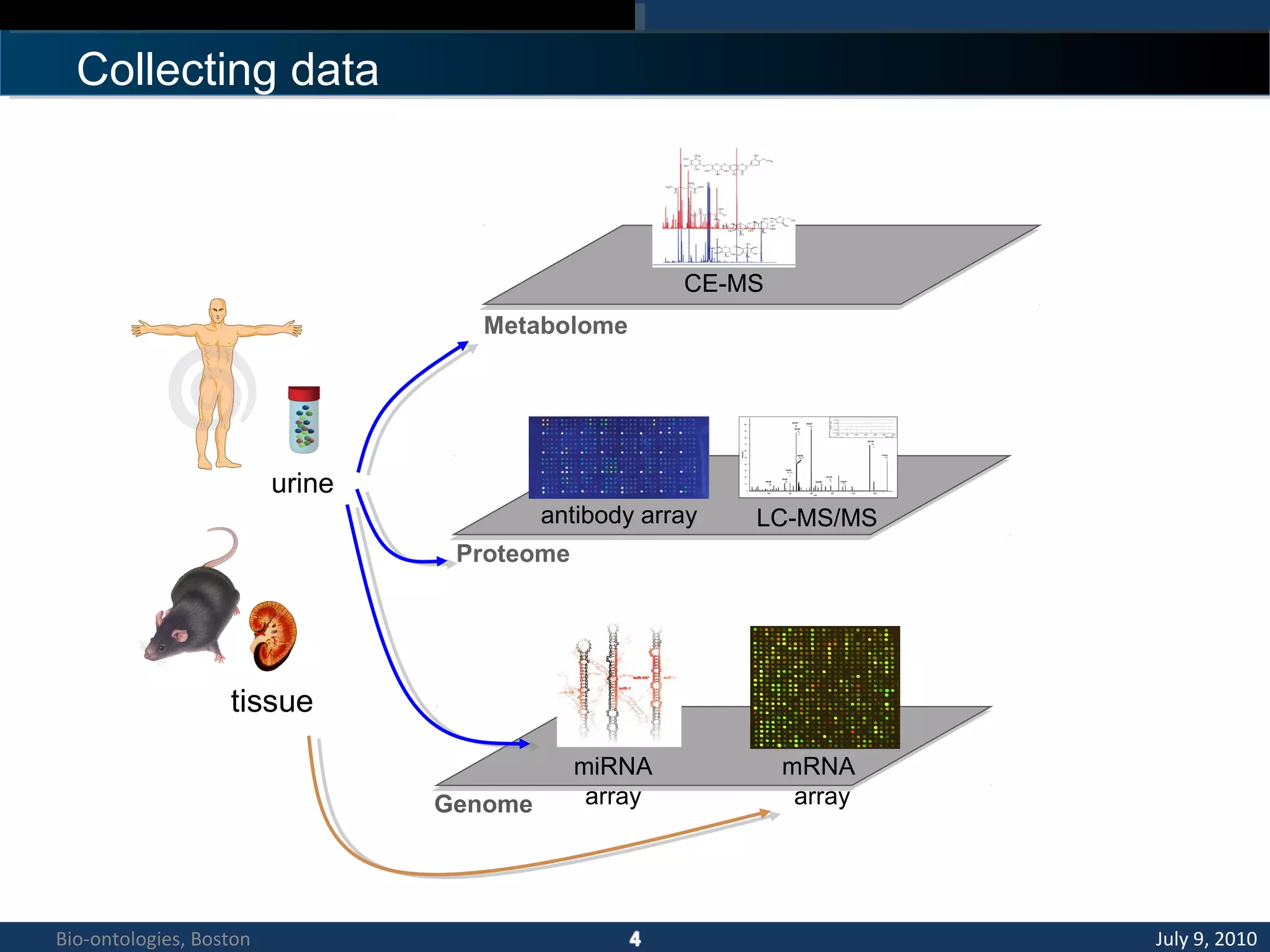
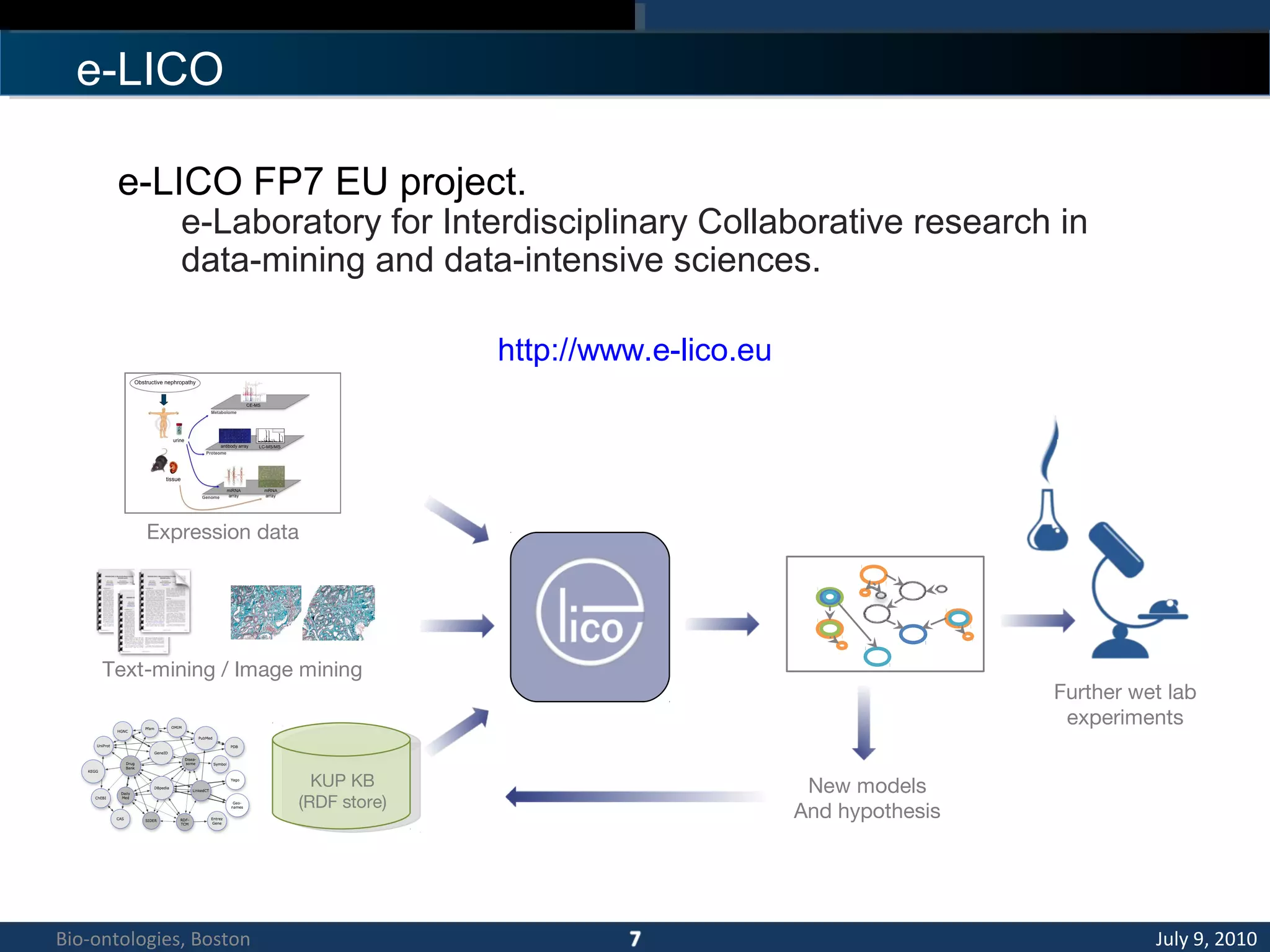
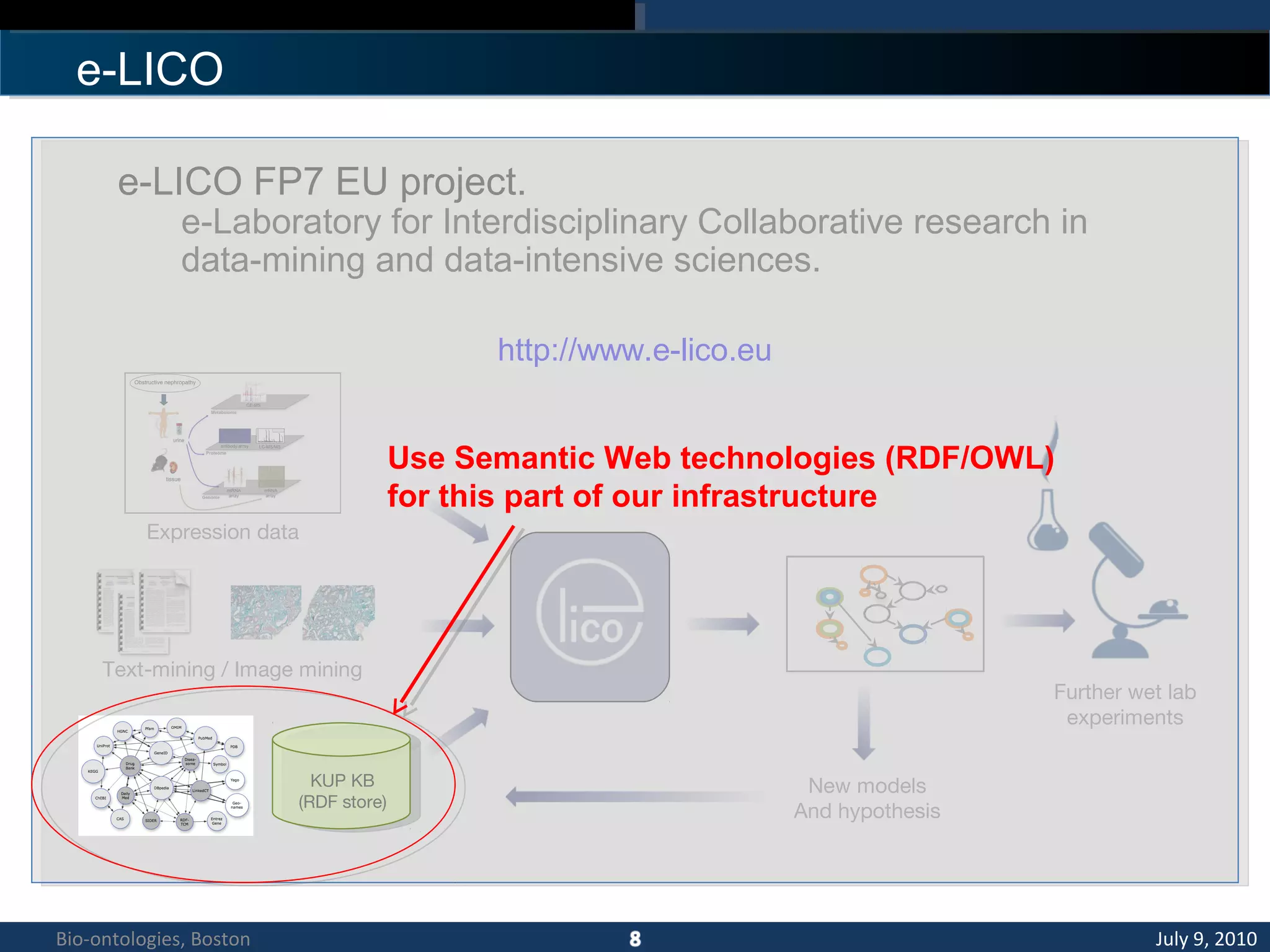
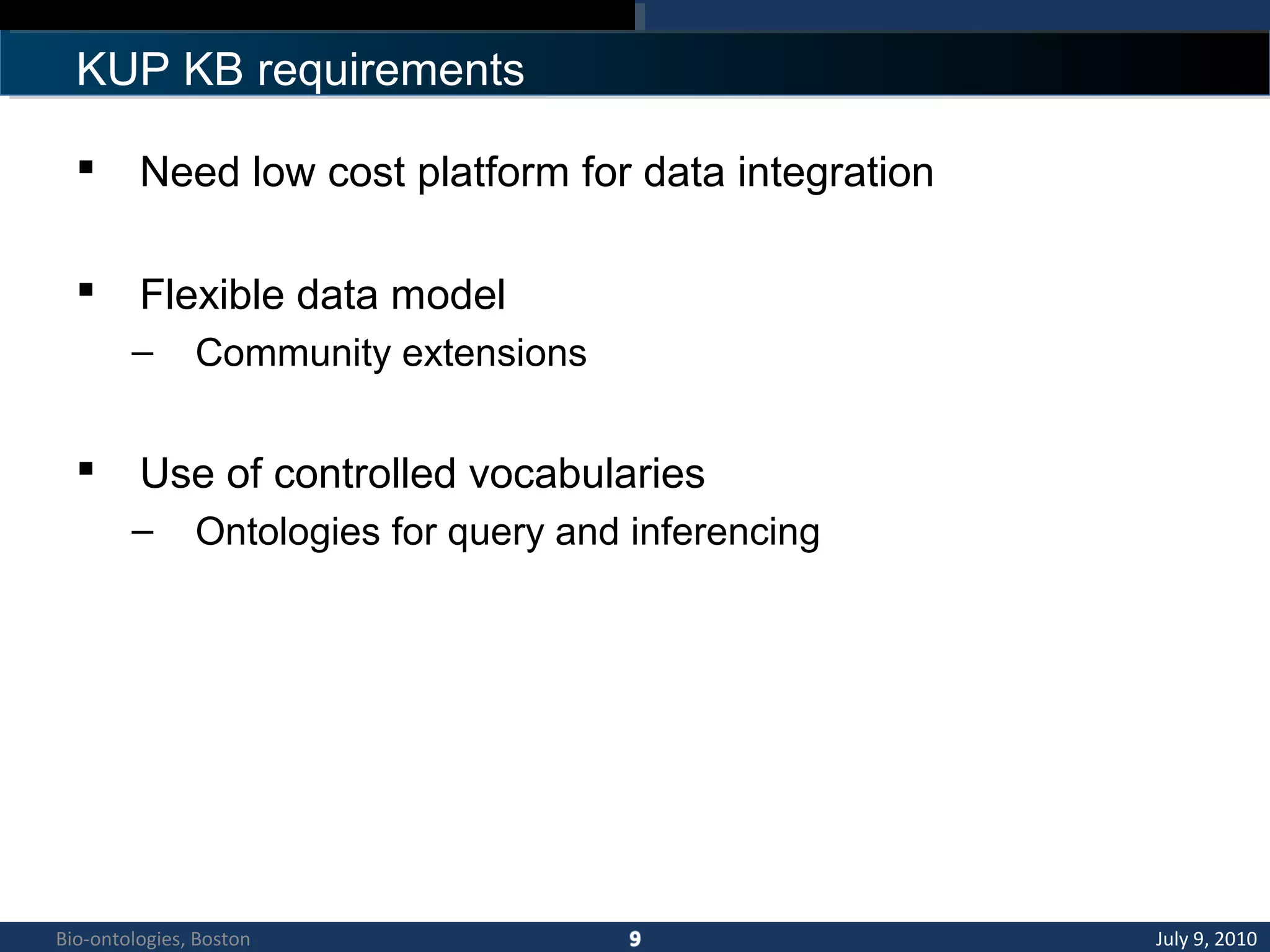
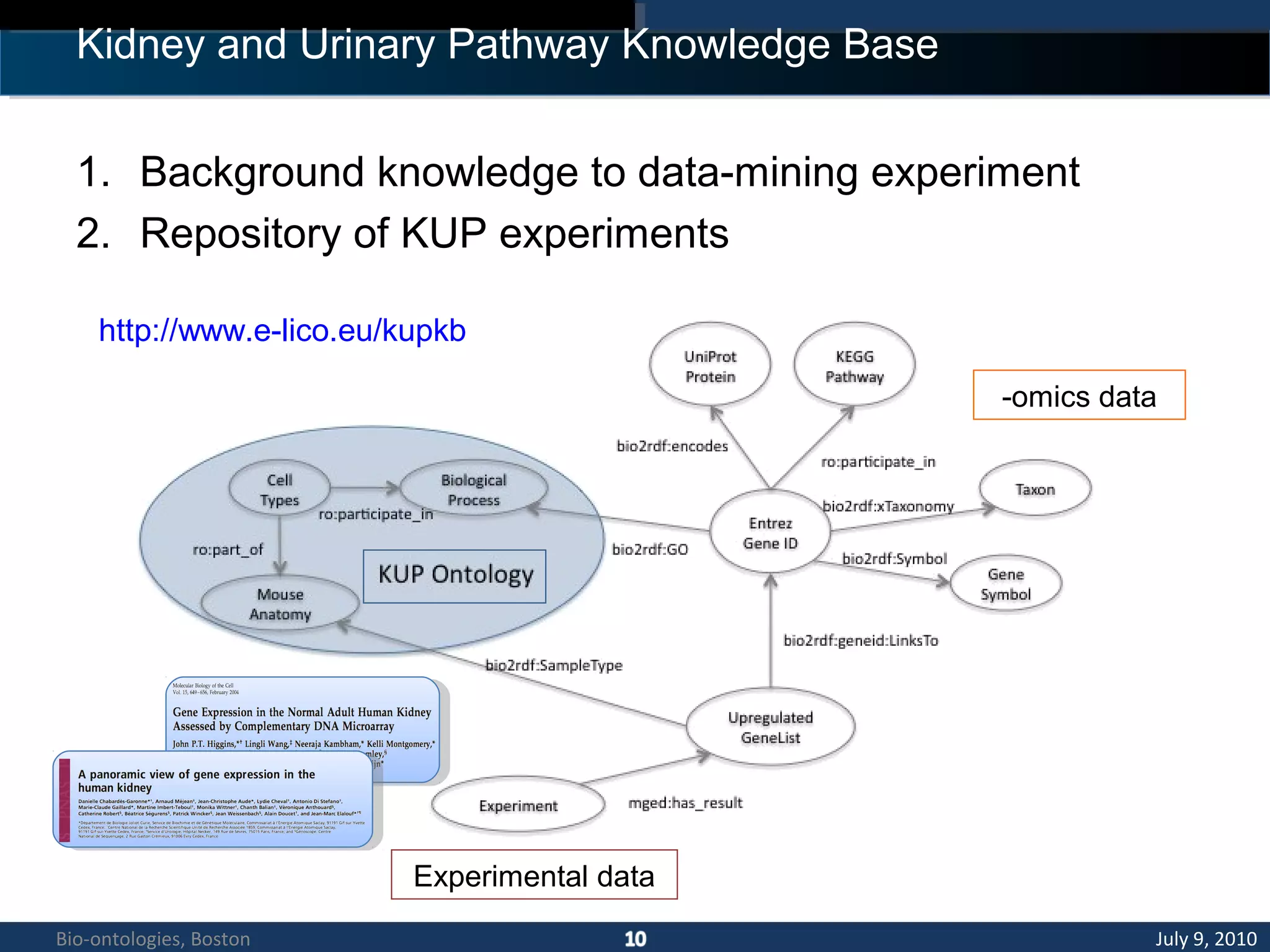
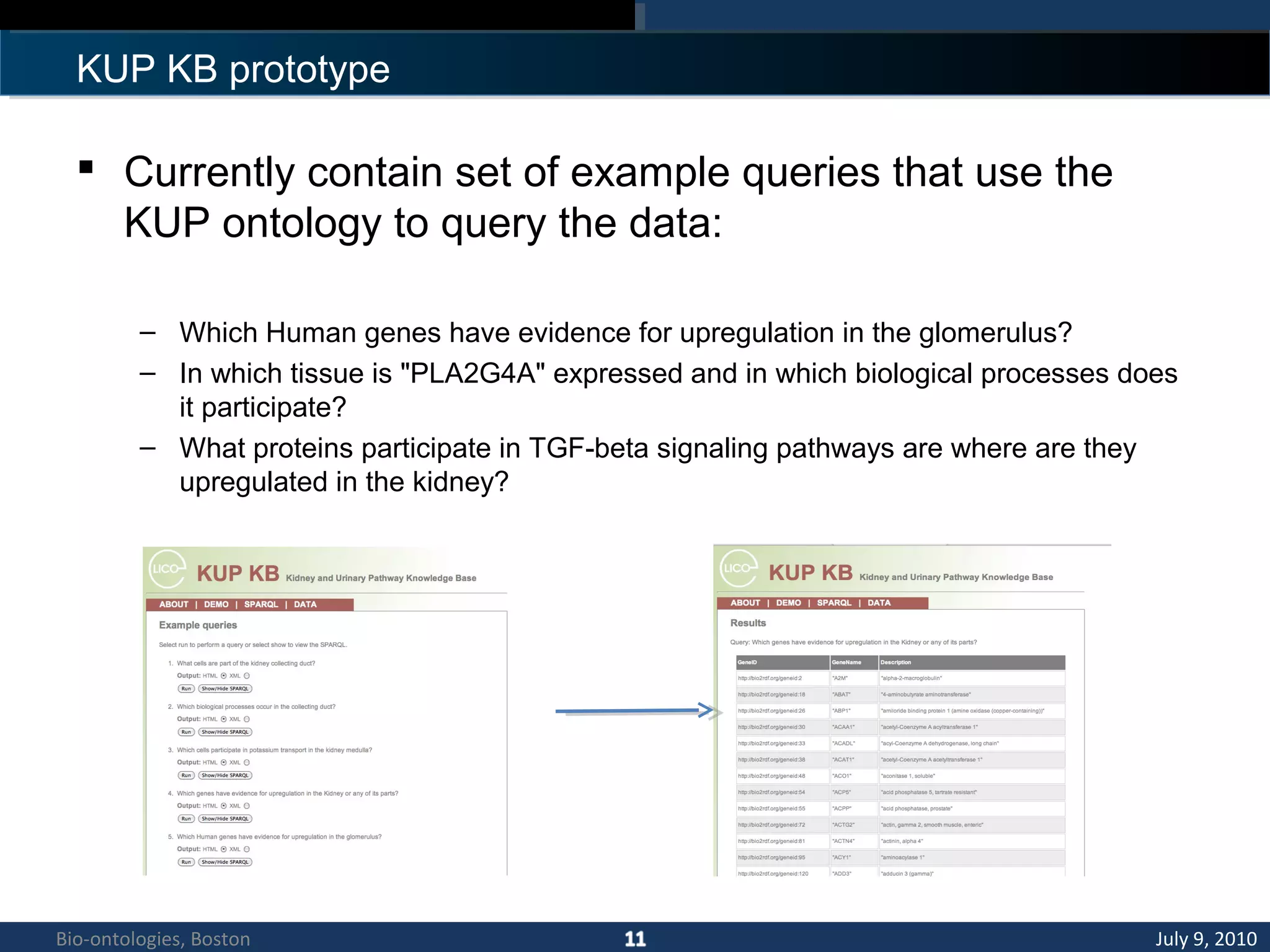
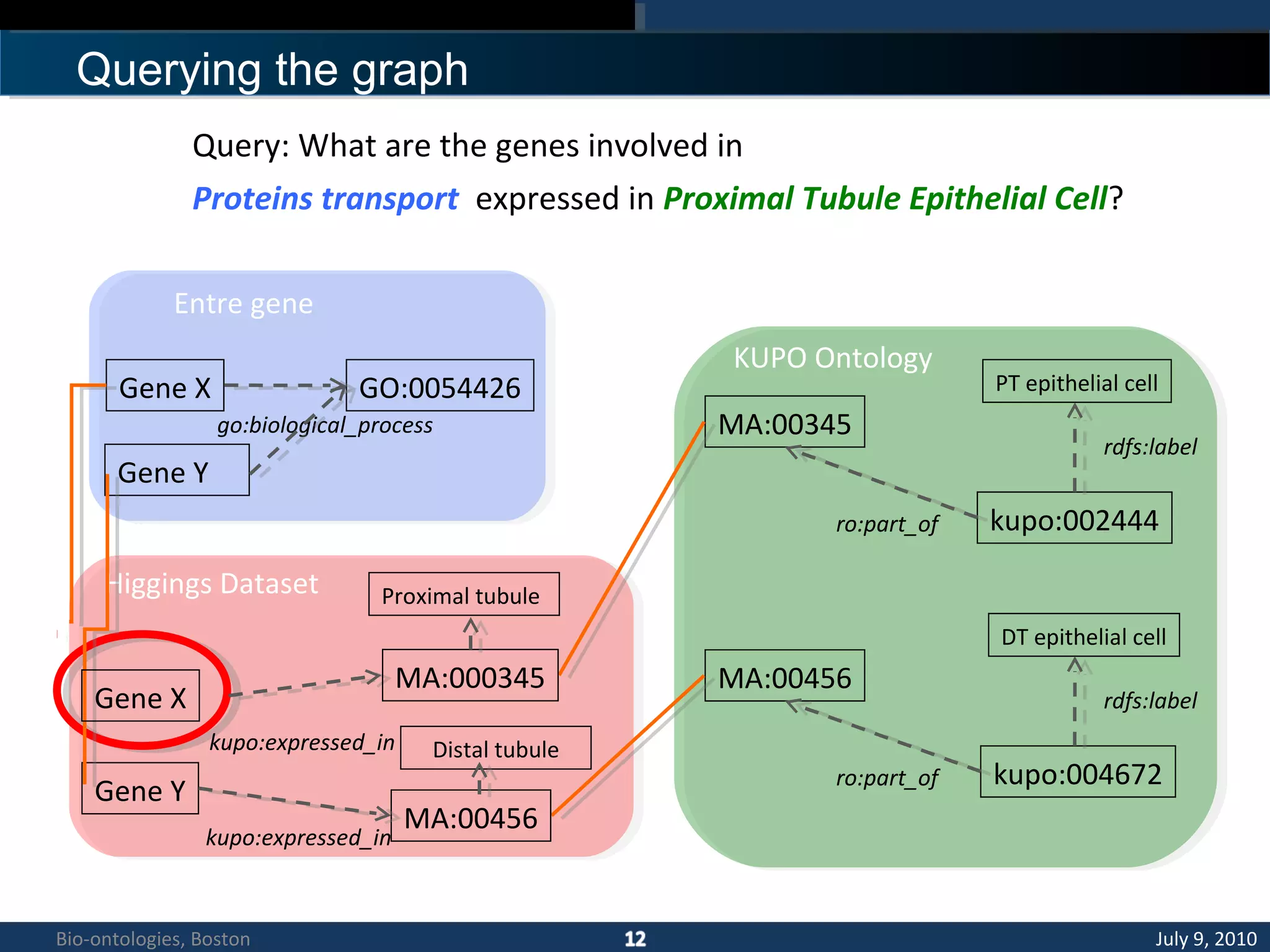
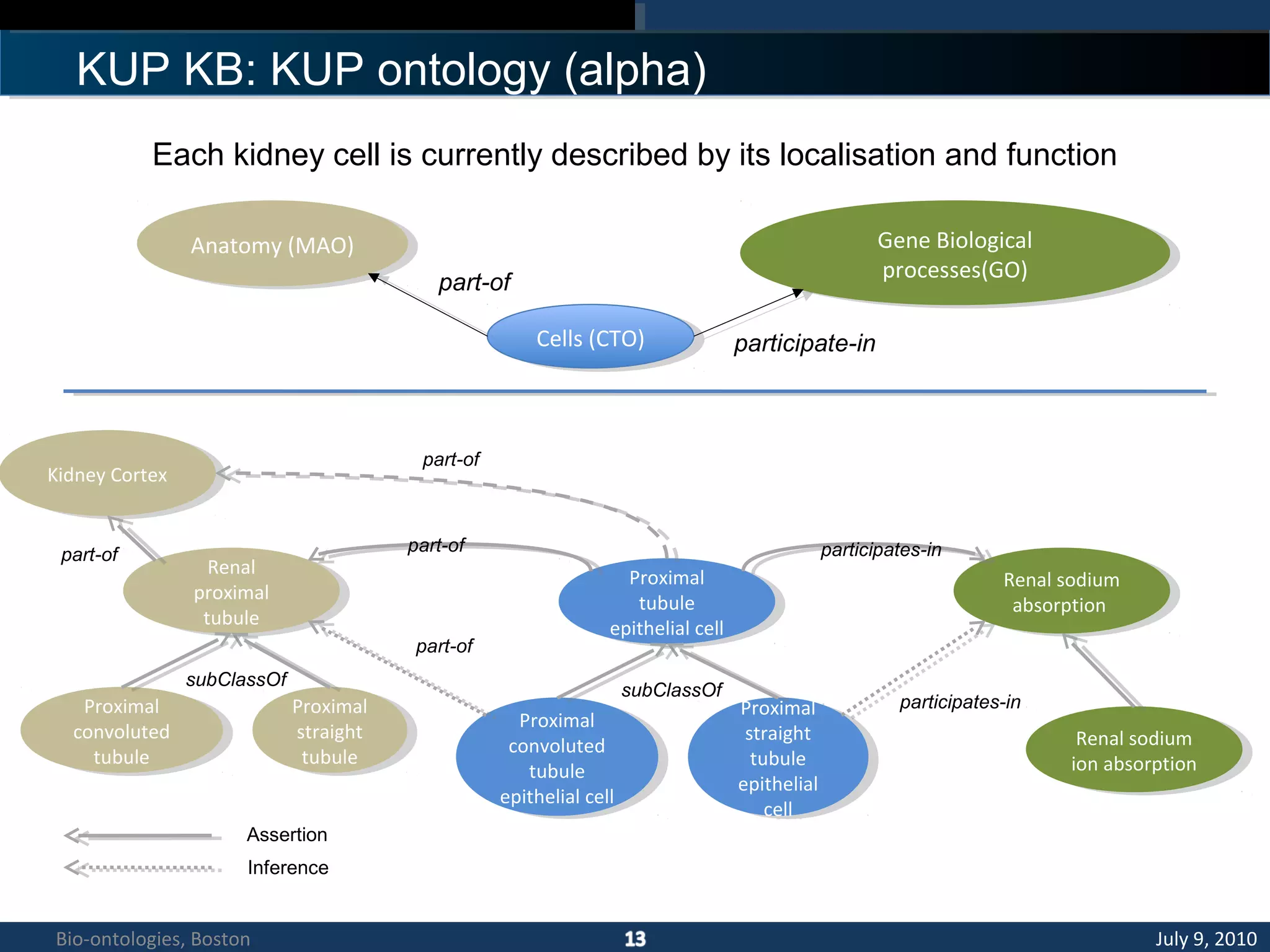
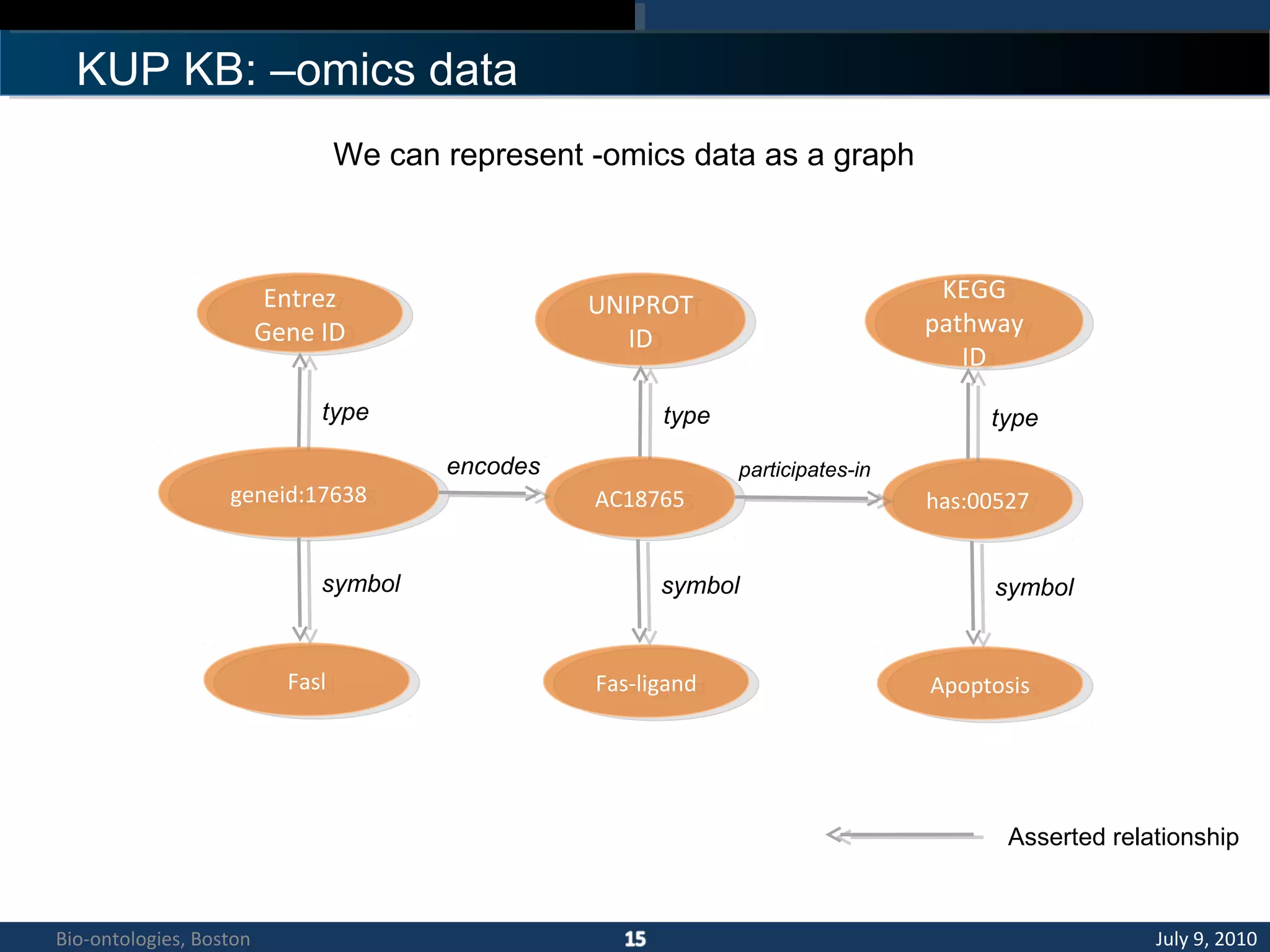
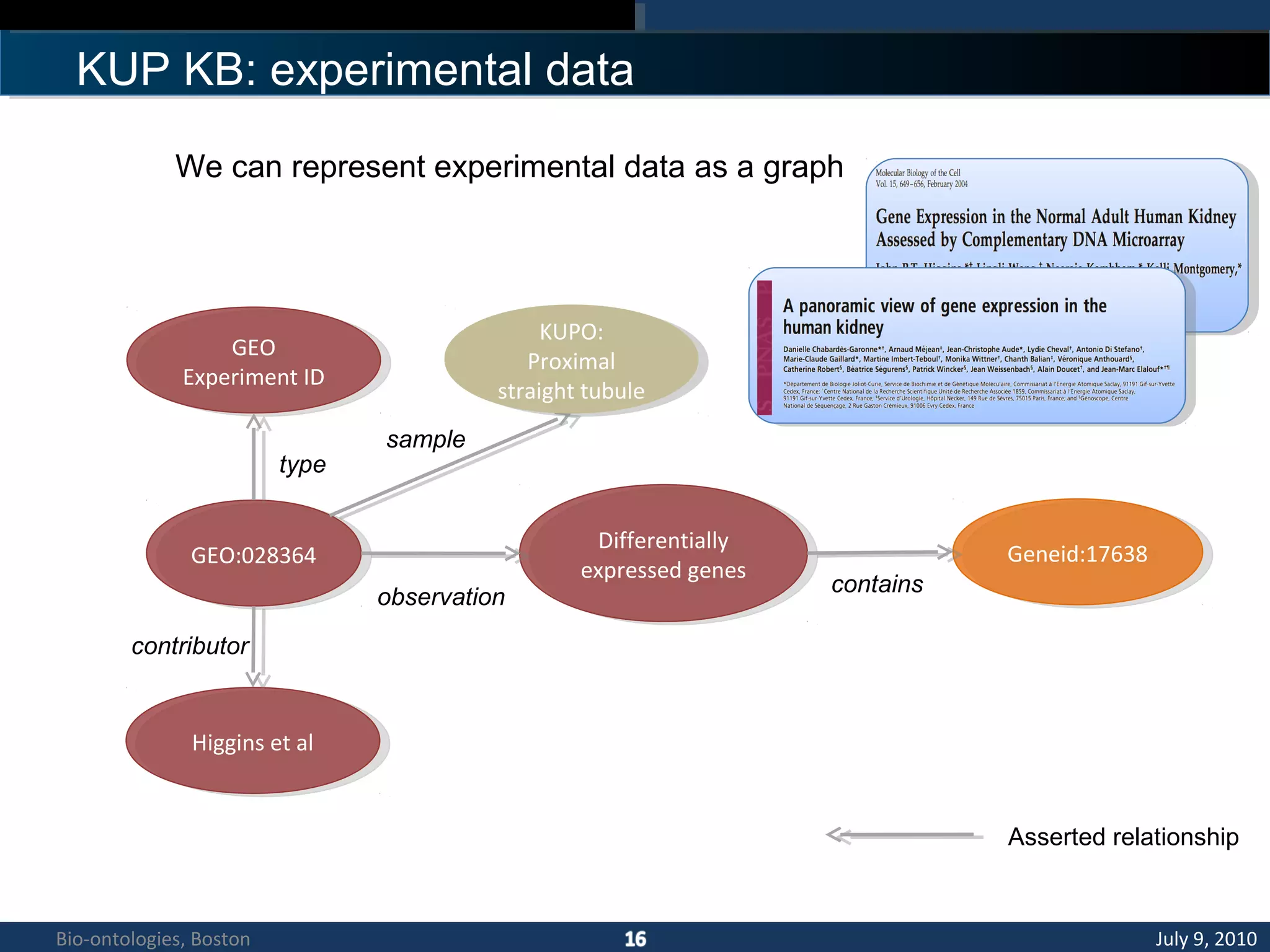
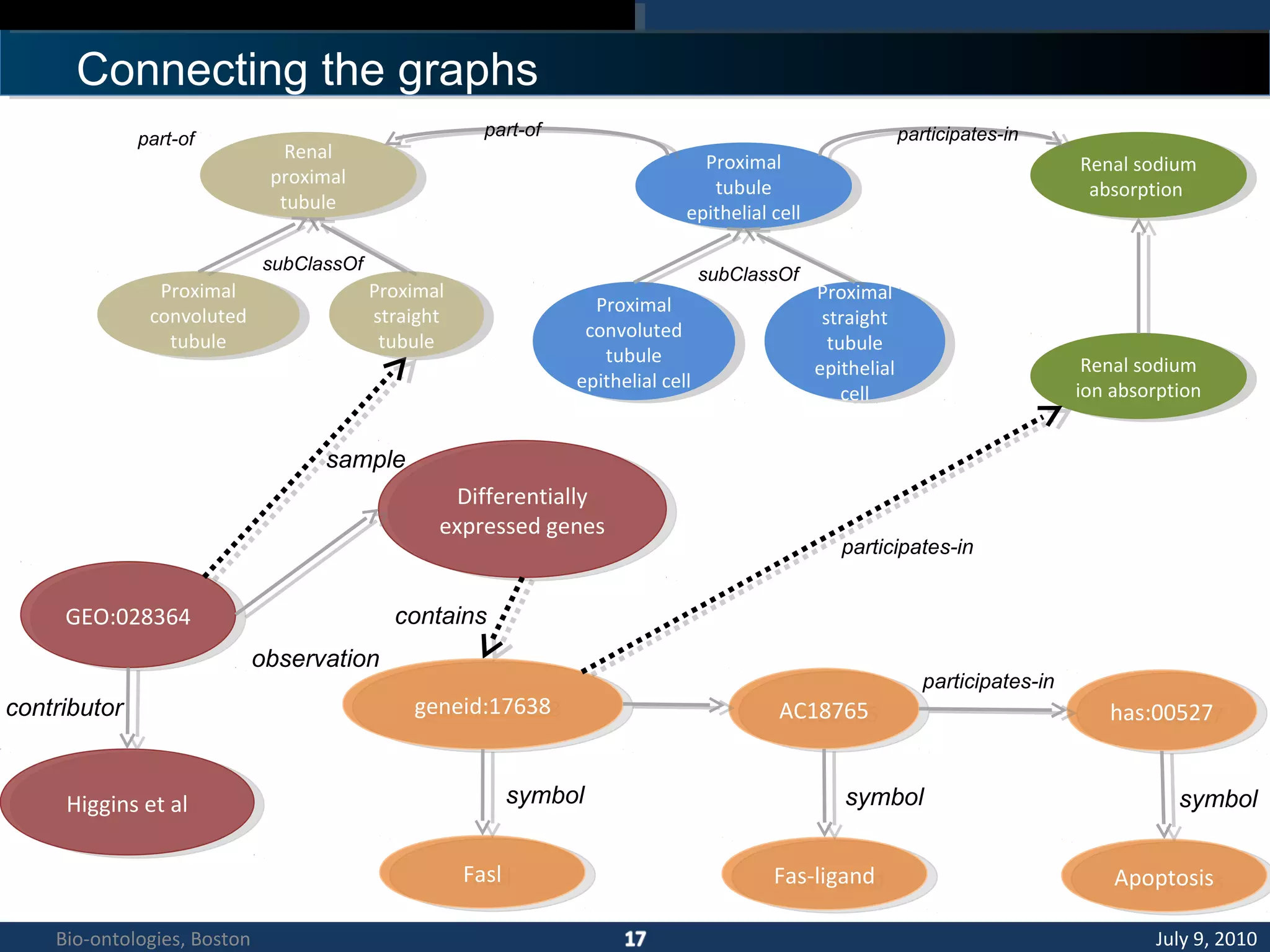
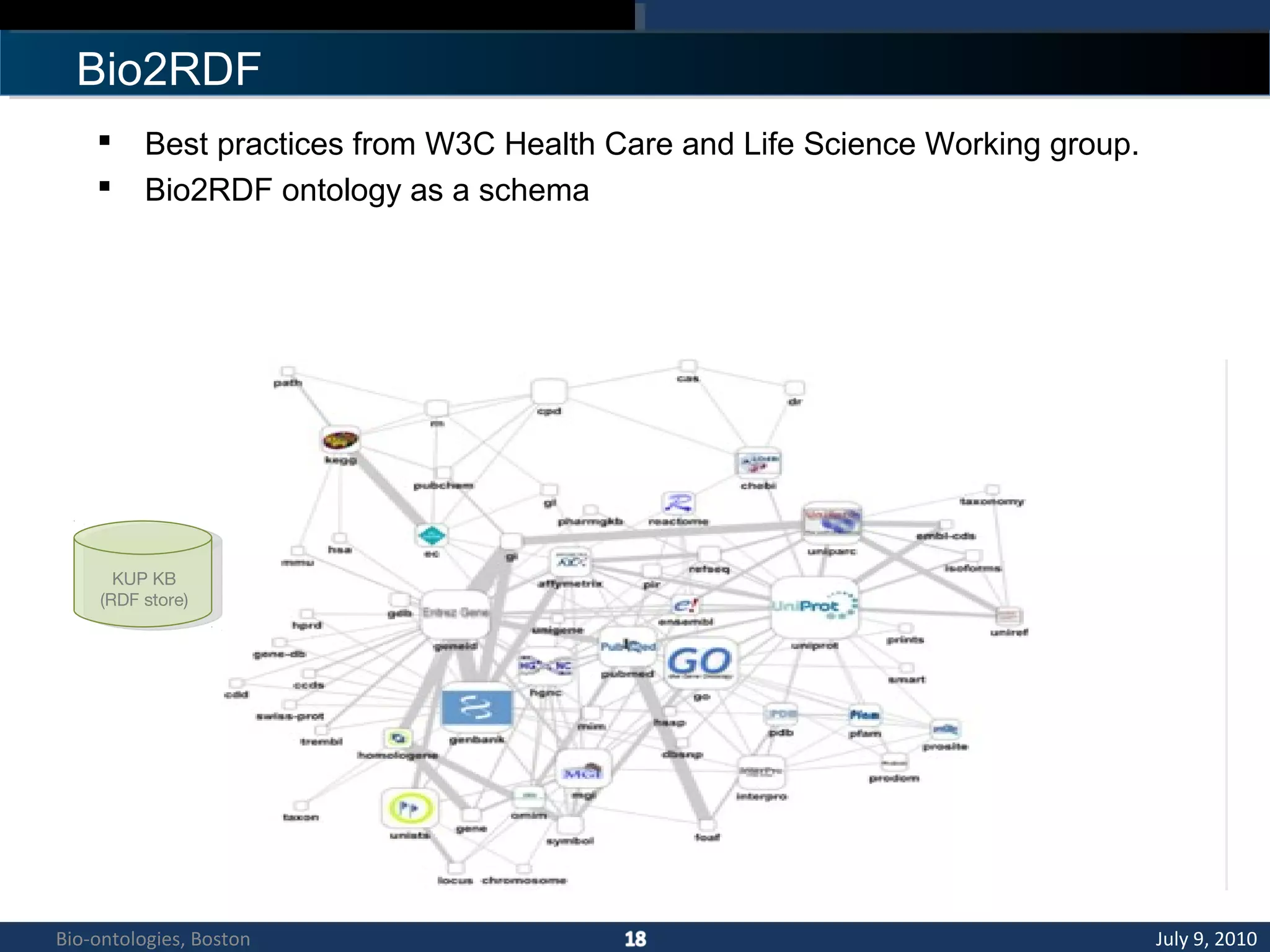
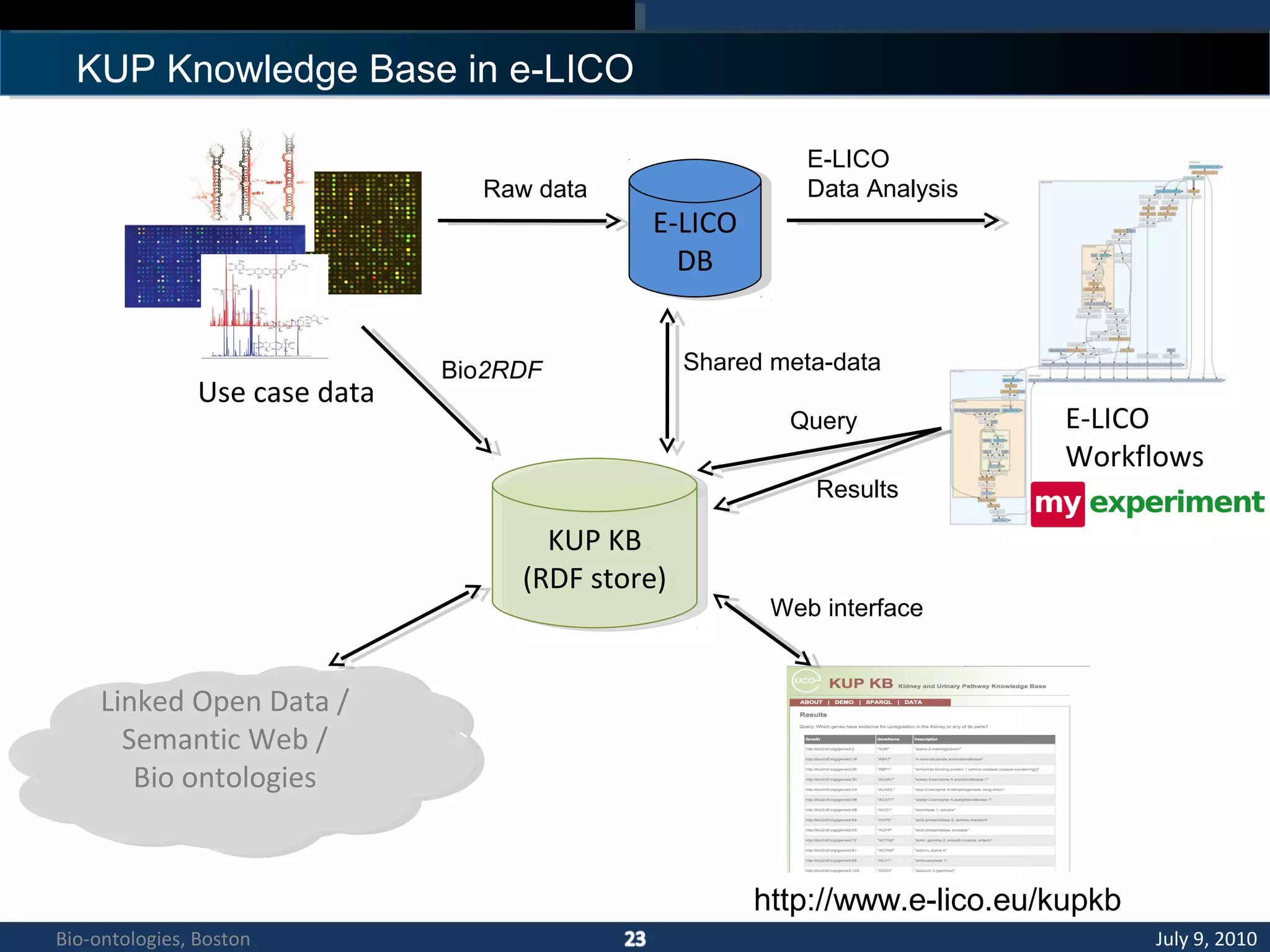
1. The document describes a kidney and urinary pathway knowledge base (KUP KB) that integrates various types of biological data using semantic web technologies.
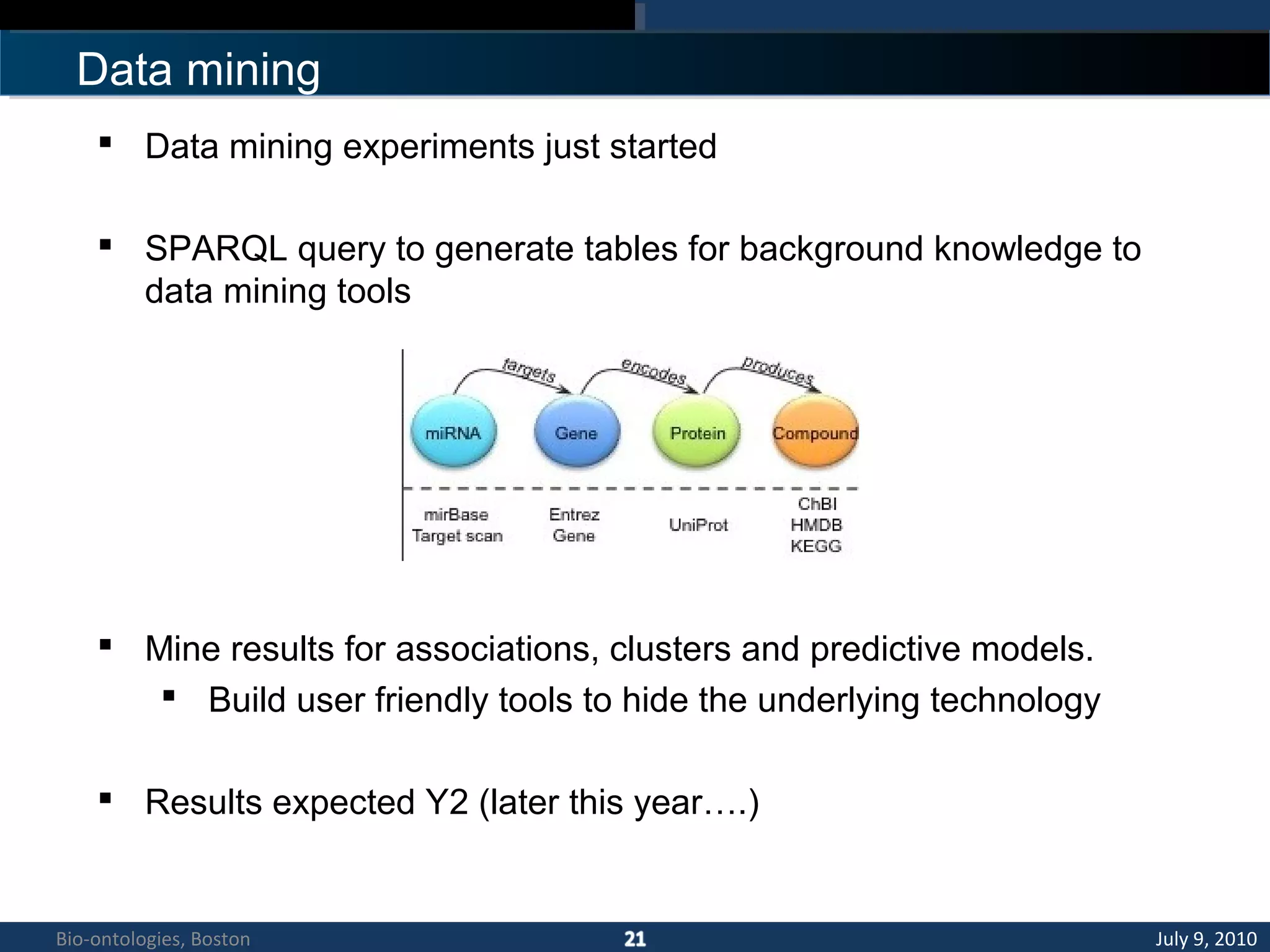
2. The KUP KB aims to provide background knowledge for data mining experiments and act as a collaborative repository for kidney and urinary pathway data.
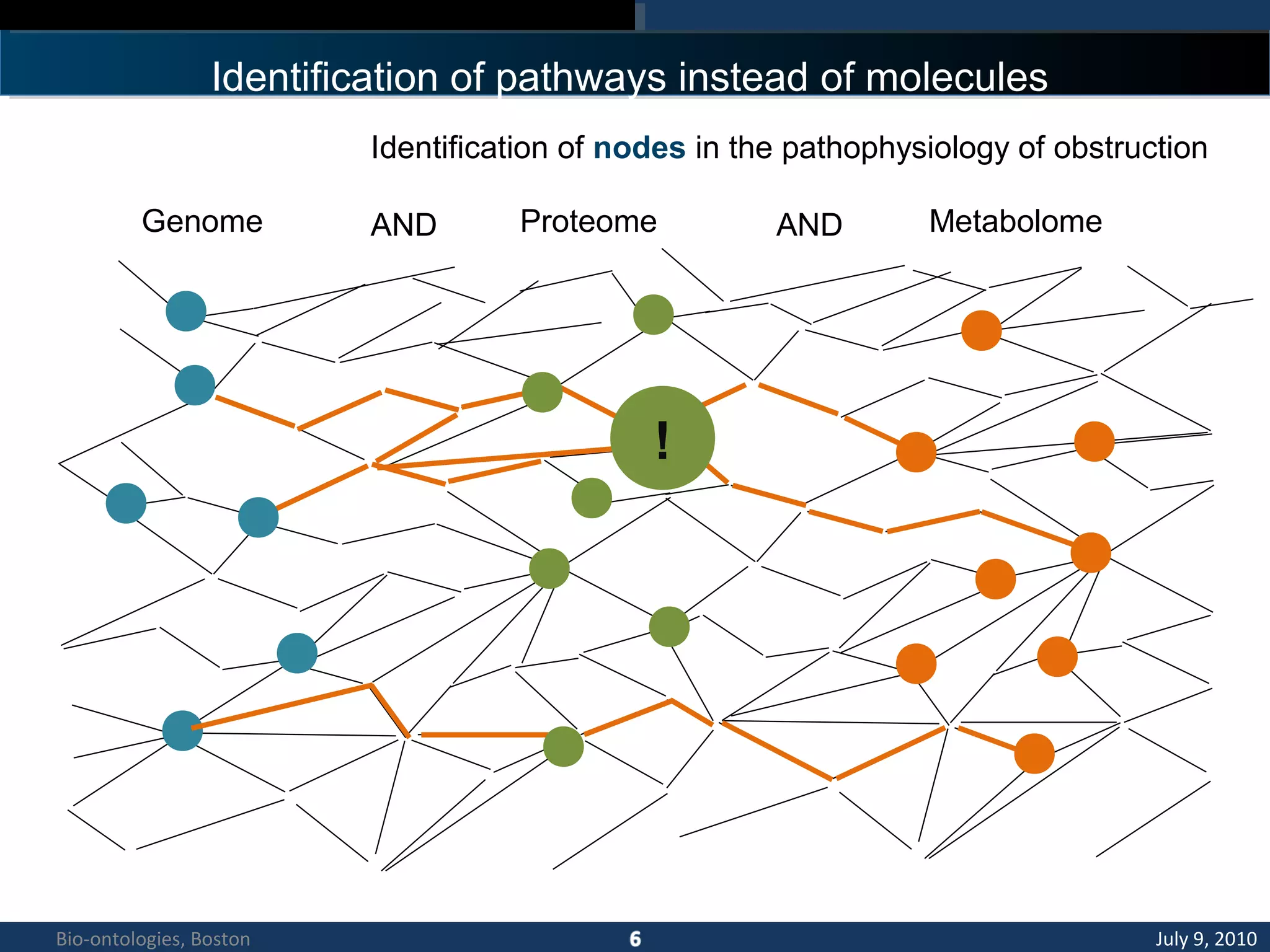
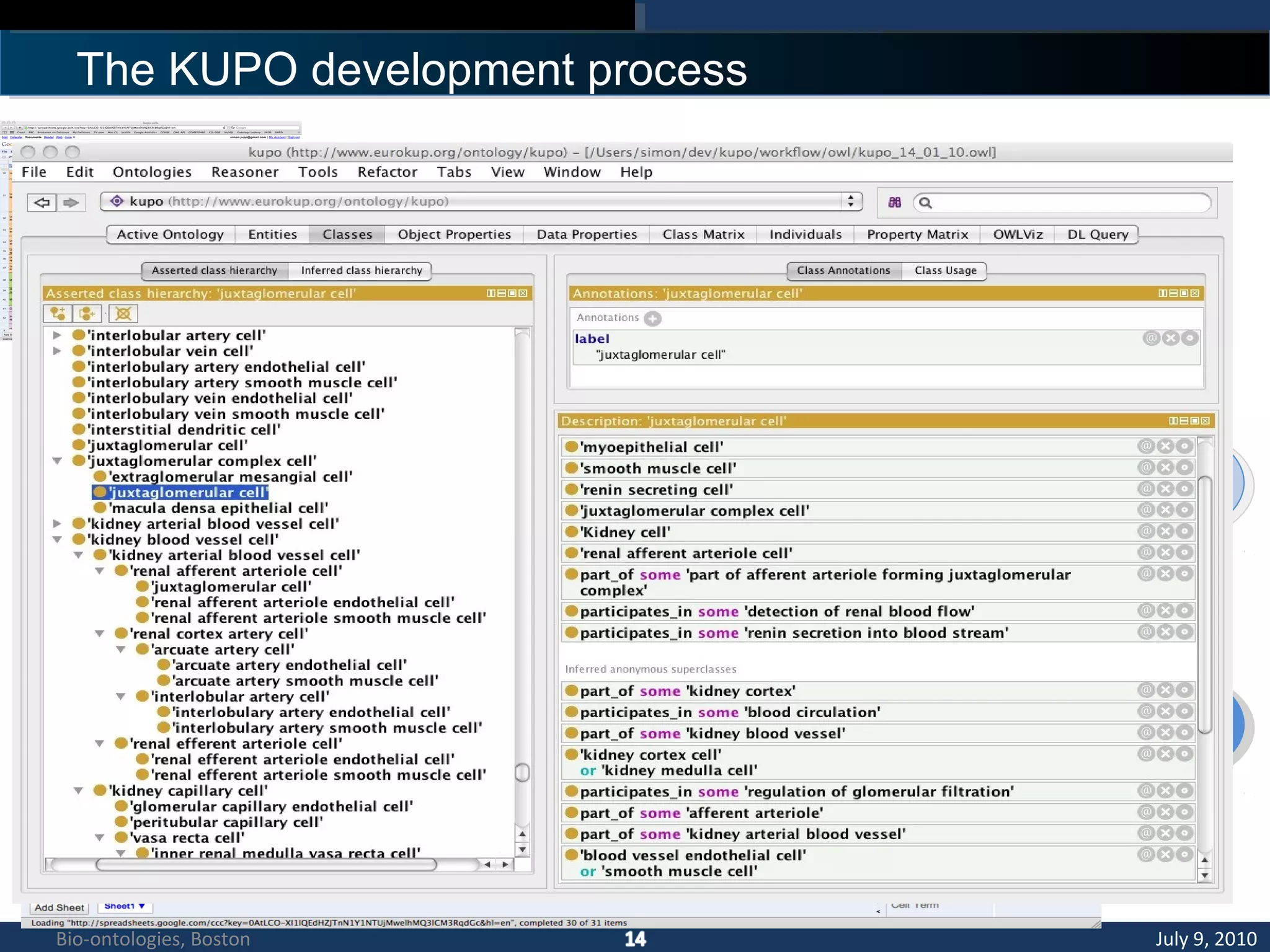
3. An ontology is being developed to represent the kidney and urinary pathway domain and enable querying across the integrated data sources in the KUP KB.