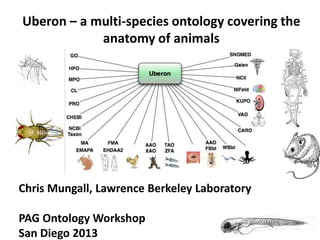
Uberon - A multi-species anatomy ontology covering animal anatomy
- 1. Uberon – a multi-species ontology covering the anatomy of animals Chris Mungall, Lawrence Berkeley Laboratory PAG Ontology Workshop San Diego 2013
- 2. Anatomical ontologies are vital for plant and animal genomics • High throughput transcriptomic and phenomic data • Need to analyze within and across species
- 3. Most animal anatomy ontologies are species specific • Mouse: – MA (adult) – EMAP / EMAPA (embryonic) • Human – FMA (adult) – EHDAA2 (CS1-CS20) • Amphibian – XAO (Xenopus) • Fish – ZFA – (Zebrafish) • Nematode – WBbt – (C elegans) • Arthropod – FBbt (Drosophila)
- 4. Ontologies built for one species will not work for others http://ccm.ucdavis.edu/bcancercd/22/mouse_figure.html http://fme.biostr.washington.edu:8080/FME/index.html
- 5. Uberon: a multi-species anatomy ontology • Contents: – Over 8,000 classes (terms) – Multiple relationships, including subclass, part-of and develops-from • Scope: metazoa (animals) – Current focus is chordates – Federated approach for other taxa • Uberon classes are generic / species neutral – ‘mammary gland’: you can use this class for any mammal! – ‘lung’: you can use this class for any vertebrate (that has lungs) Mungall, C. J., Torniai, C., Gkoutos, G. V., Lewis, S. E., & Haendel, M. A. (2012). Uberon, an integrative multi-species anatomy ontology. Genome Biology, 13(1), R5. doi:10.1186/gb- 2012-13-1-r5 http://genomebiology.com/2012/13/1/R5
- 6. is_a (SubClassOf) anatomical part_of structure develops_from capable_of is_a (taxon equivalent) endoderm only_in_taxon organ part foregut swim bladder organ endoderm of forgut NCBITaxon: respiration organ Actinopterygii respiratory primordium GO: respiratory gaseous exchange pulmonary acinus alveolus lung lung primordium NCBITaxon: Mammalia alveolus of lung alveolar sac lung bud FMA: pulmonary FMA:lung MA:lung alveolus EHDAA: MA:lung alveolus lung bud
- 7. Bridging anatomy ontologies SNOMED Uberon NCIt ZFA EMAPA EHDAA2 MA FMA CJ Mungall, C Torniai, GV Gkoutos, SE Lewis, MA Haendel. Uberon, an integrative multi-species anatomy ontology. Genome biology 13 (1), R5
- 8. Inter-ontology links UBERON: respiratory system capable_of GO: respiratory gaseous exchange part_of UBERON: respiratory airway is_a only_in_taxon UBERON: trachea Vertebrata CL: epithelial cell part_of is_a CL: tracheal epithelial cell
- 9. Uberon PURLs Permanent URL Humans see HTML view
- 10. Uberon PURLs Machines see this
- 11. Different versions for different purposes ontology contents basic simple relationships http://purl.obolibrary.org/obo/uberon/basic.owl uberon main ontology http://purl.obolibrary.org/obo/uberon.owl merged main ontology + links to GO, CL, NCBITaxon, NBO http://purl.obolibrary.org/obo/uberon/merged.owl Composite- Uberon plus species-specific ontology classes merged in metazoan http://purl.obolibrary.org/obo/uberon/composite-metazoan.owl Formats: http://uberon.org • OBO-Format • OWL
- 12. Efficient Ontology Development Text processing Ontology matching Wikipedia extraction Automated Reasoning Expert Curation • Automated QC checks in Multiple knowledge Jenkins pipeline[*] acquisution tools • Automated hierarchy building Content meetings using OWL reasoners Expert interviews * http://bio-ontologies.knowledgeblog.org/405 http://owltools.googlecode.com/
- 13. Applications • Phenomics: comparing phenotypes across species • Transcriptomics: gene expression • Systematics and evolutionary biology
- 14. Finding disease models • Washington, N. L., Haendel, M. A., Mungall, C. J., Ashburner, M., Westerfield, M., & Lewis, S. E. (2009). Linking Human Diseases to Animal Models Using Ontology-Based Phenotype Annotation. PLoS Biol, 7(11). Public Library of Science. doi:10.1371/journal.pbio.1000247 • Chen, C.-K., Mungall, C. J., Gkoutos, G. V., Doelken, S. C., Koehler, S., Ruef, B. J., Smith, C., et al. (2012). MouseFinder: Candidate disease genes from mouse phenotype data. Human mutation, 33(5), 858-66. doi:10.1002/humu.22051
- 15. Uberon and FANTOM5 Kenneth Baillie, FANTOM5 consortium
- 16. http://bgee.unil.ch Niknejad, A., Comte, A., Parmentier, G., Roux, J., Frederic, B., & Robinson-rechavi, M. (2011). vHOG , a multi-species vertebrate ontology of homologous or- gans groups. Ecology, 1-5.
- 18. LAMHDI: Linking across models for human disease initiative • Integrating multiple data sources – OMIM, OMIA (Online Mendelian Inheritance in Animals), model organisms, phenotype and trait data, etc – Annotating using ontology terms • Providing novel semantic linking and navigation methods http://lamhdi.org (early prototype) https://neuinfo.org/mynif/ (search)
- 19. Beyond the core model organisms • Phase 1 development: 2009-2011 – Core model organism and human anatomy • Phase 2 development: 2012 – Vertebrate skeletal system • Collaboration with phenoscape • Incorporate TAO (teleost), AAO (amphibian), VSAO (vertebrate skeleton) • Phase 3 development: 2013- – Other taxa • E.g – Ruminants – Aves
- 20. Conclusions • Most animal anatomy ontologies are specialized for core model organisms or humans • Uberon bridges across species-specific anatomical ontologies and is a complete ontology in its own right • Uberon is richly connected with other orthogonal ontologies and is being used for annotation in a variety of projects
- 21. Get involved • Tracker – http://purl.obolibrary.org/obo/uberon/tracker • Mailing list – https://lists.sourceforge.net/lists/listinfo/obo- anatomy • Website – http://uberon.org • Data (coming soon) – http://bgee.unil.ch – http://kb.phenoscape.org – http://lamhdi.org
- 22. Acknowledgments Uberon Developers Contributors Contributors • Carlo Torniai (eagle i) • Erik Segerdell (XAO / • Alex Dececchi Phenotype RCN) • Nizar Ibrahim • George Gkoutos (NBO) • Paul Sereno (Phenoscape) • Melissa Haendel • Jonathan Bard (EHDAA2) • Monte Westerfield (ZFA) • Terry Meehan (CL) • Cynthia Smith (MP) • Alex Diehl (CL) • Maryanne Martone (NIF) Applications and • Bill Bug (NIF) software development • Terry Hayamizu (MA/CL) • Aurelie Comte (Bgee) • Frederic Bastian • Yvonne Bradford (ZFA) • Anna Niknejad (Bgee) • Heiko Dietze • Ceri van Slyke (ZFA) • Marc Robinson-Rechavi • Jim Balhoff • David Hill (GO) (Bgee) • Paula Mabee • David Osumi Sutherland • Robert Druzinsky (FEED) (FBbt/CARO) • Brian K Hall (neural crest) • Suzanna Lewis • Paul Schofield (MPATH) • Sarah Whitcher Kansa • Ann Maglia (AAO) • Wasilla Dahdul • Paul Sereno (Phenoscape) (TAO/VAO) • David Blackburn (Phenoscape)
Editor's Notes
- Images: Seth Ruffins
- UBERON uses GO or other external ontologies for logicaldefinitions (e.g. chemosensory organ, respiration organ, reproductivesystem -- GO; smooth muscle tissue - CL)
