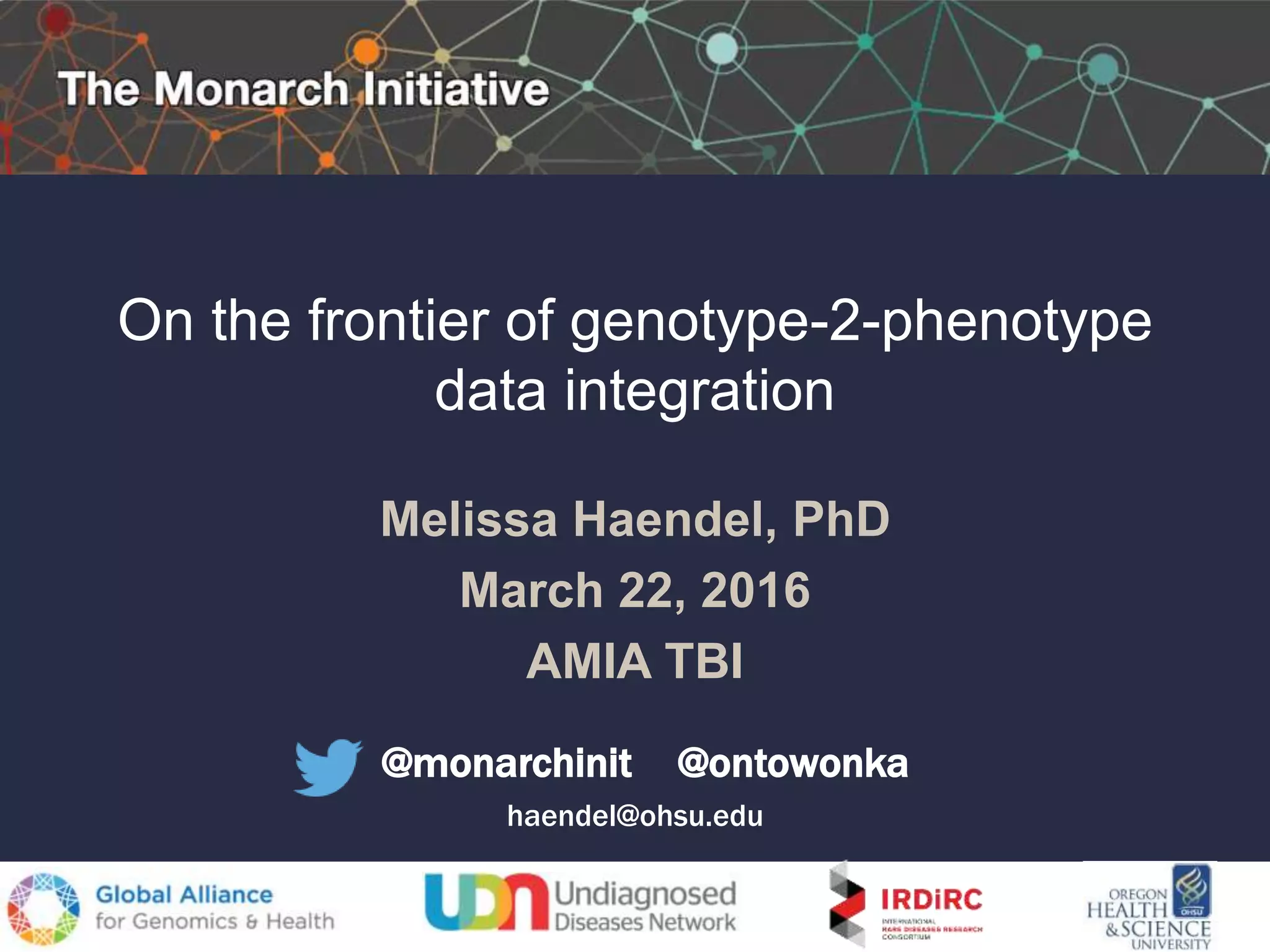
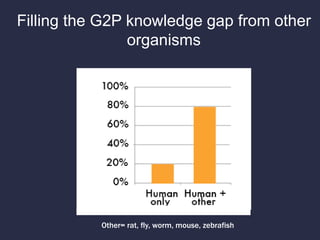
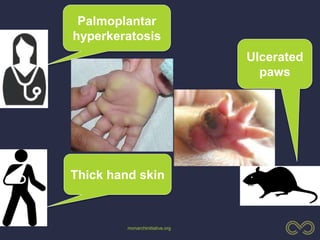
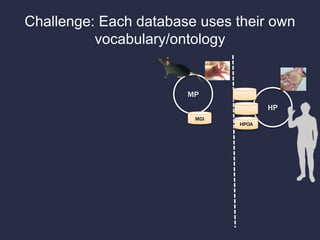
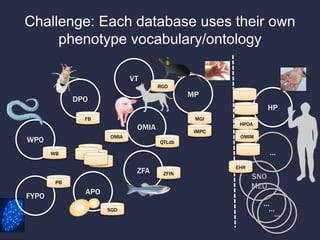
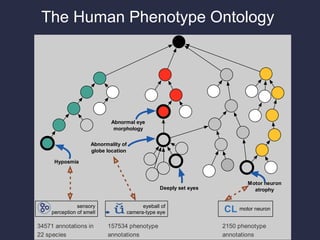
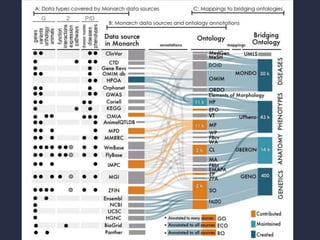
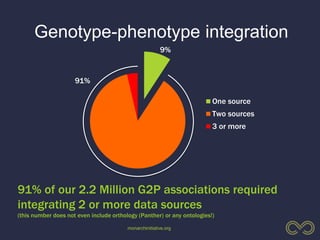
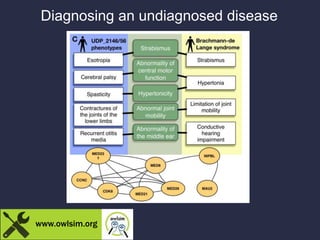
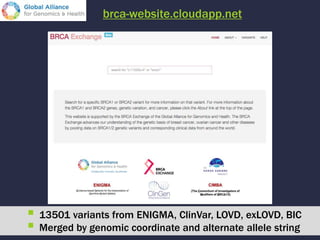
The document discusses the challenges of integrating genotype-phenotype (G2P) data across various organisms using different vocabularies and ontologies. It emphasizes the necessity for a standardized phenotypic exchange and computable models to improve data sharing and variant interpretation, ultimately aiding in disease diagnosis and research. Acknowledgments are given to various contributors and funding sources involved in this initiative.