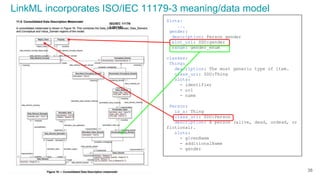
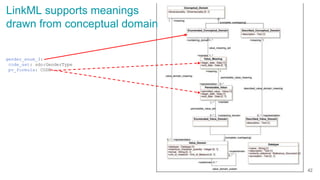
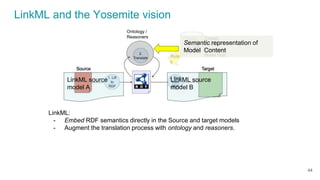
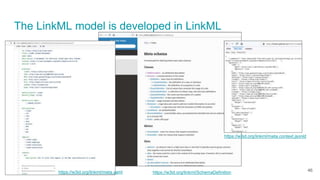
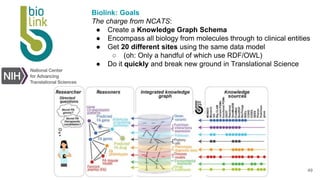
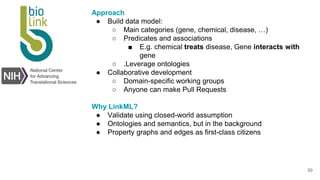
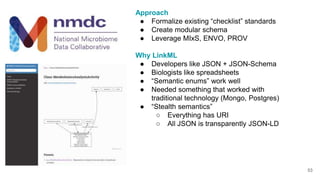
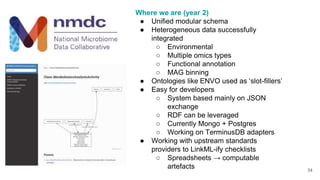
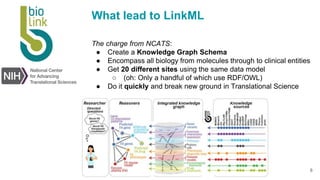
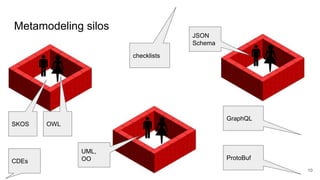
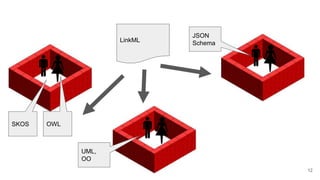
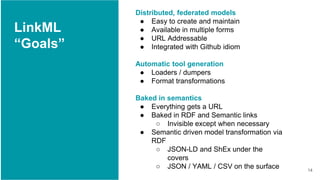
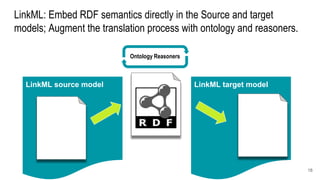
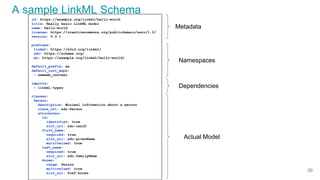
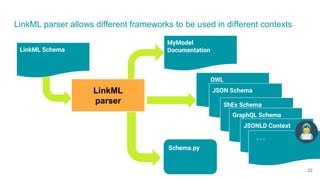
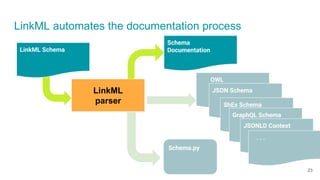
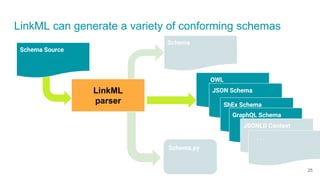
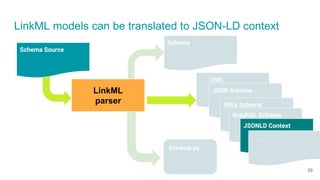
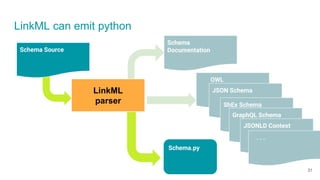
LinkML is a modeling language for building semantic models that can be used to represent biomedical and other scientific knowledge. It allows generating various schemas and representations like OWL, JSON Schema, GraphQL from a single semantic model specification. The key advantages of LinkML include simplicity through YAML files, ability to represent models in multiple forms like JSON, RDF, and property graphs, and "stealth semantics" where semantic representations like RDF are generated behind the scenes.








![BASE <https://example.org/linkml/hello-world/>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX sdo: <https://schema.org/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
<String> xsd:string
<Person> CLOSED {
( $<Person_tes> ( sdo:givenName @<String> + ;
sdo:familyName @<String> ;
foaf:knows @<Person> *
) ;
rdf:type [ sdo:Person ]
)
}
"$id": "https://example.org/linkml/person",
"$schema": "http://json-schema.org/draft-07/schema#",
"definitions": {
"Person": {
"additionalProperties": false,
"description": "Minimal information about a person",
"properties": {
"first_name": {
"items": {
"type": "string"
},
"type": "array"
},
"id": {
"type": "string"
},
...
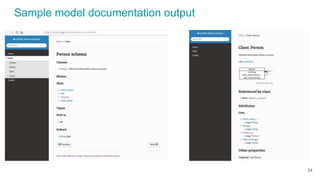
Shape Expressions (ShEx) Schema
JSON Schema
type Person
{
id: String!
firstName: [String]!
lastName: String!
knows: [Person]
}
Graphql Schema
Sample LinkML generated schemas
26](https://image.slidesharecdn.com/linkmlyosemitepresentation-211018192042/85/LinkML-presentation-to-Yosemite-Group-26-320.jpg)
![# Types
class String(str):
type_class_uri = XSD.string
type_class_curie = "xsd:string"
type_name = "string"
type_model_uri = EX.String
@dataclass
class Person(YAMLRoot):
"""
Minimal information about a person
"""
id: Union[str, PersonId] = None
first_name: Union[str, List[str]] = None
last_name: str = None
knows: Optional[Union[Union[str, PersonId], List[Union[str, PersonId]]]] = empty_list()
def __post_init__(self, *_: List[str], **kwargs: Dict[str, Any]):
if self.id is None:
raise ValueError("id must be supplied")
if not isinstance(self.id, PersonId):
self.id = PersonId(self.id)
if self.first_name is None:
raise ValueError("first_name must be supplied")
elif not isinstance(self.first_name, list):
self.first_name = [self.first_name]
elif len(self.first_name) == 0:
raise ValueError(f"first_name must be a non-empty list")
self.first_name = [v if isinstance(v, str) else str(v) for v in self.first_name]
...
from examples.basic import Person
sam = Person("1172438", first_name=["Samual", "J"],last_name="Snooter")
print(sam)
Person(id='1172438', first_name=['Samual', 'J'], last_name='Snooter',
knows=[])
fred = Person("a117", first_name="John")
...
ValueError: last_name must be supplied
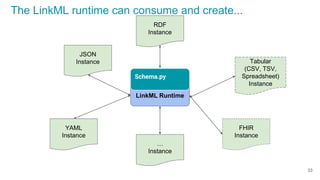
Using python code emitted by LinkML
32](https://image.slidesharecdn.com/linkmlyosemitepresentation-211018192042/85/LinkML-presentation-to-Yosemite-Group-32-320.jpg)
![from examples.basic import Person
from linkml.dumpers import json_dumper, rdf_dumper
sam = Person("1172438", first_name=["Samual", "J"], last_name="Snooter")
ann = Person("17a3923", first_name="Jill", last_name="Jones", knows=[sam.id])
print(json_dumper.dumps(ann))
print(yaml_dumper.dumps(ann))
print(rdf_dumper.dumps(ann, contexts="../examples/jsonld/basic.context.jsonld"))
{
"id": "17a3923",
"first_name": [
"Jill"
],
"last_name": "Jones",
"knows": [
"1172438"
],
"@type": "Person"
}
id: 17a3923
first_name:
- Jill
last_name: Jones
knows:
- '1172438'
@prefix foaf: <http://xmlns.com/foaf/0.1/> .
@prefix sdo: <https://schema.org/> .
<https://example.org/linkml/hello-world/17a3923> a sdo:Person ;
foaf:knows <https://example.org/linkml/hello-
world/1172438> ;
sdo:familyName "Jones" ;
sdo:givenName "Jill" .
python
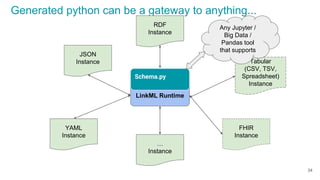
JSON output YAML output RDF output (by way of JSON-LD)
Objects can be exported as JSON, YAML, or RDF
35](https://image.slidesharecdn.com/linkmlyosemitepresentation-211018192042/85/LinkML-presentation-to-Yosemite-Group-35-320.jpg)
![from linkml.loaders import yaml_loader
fred = yaml_loader.load('input/fred.yaml', target_class=Person)
print(fred.first_name)
['Fred', 'William']
harvey = json_loader.load('https://raw.githubusercontent.com/hsolbrig/linkml-enhanced-
template/master/tests/input/harvey.json', target_class=Person)
print(harvey.last_name)
Mackerson
ann = rdf_loader.load('input/ann.xml', target_class=Person, fmt="xml")
print(ann.last_name)
Richardson
id: 118-28-3199
first_name:
- Fred
- William
last_name: Phillips
knows:
- '1172438'
- '1172438'
input/fred.yaml
Python code
{
"id": "118-78-0697",
"first_name": [
"Harvey"
],
"last_name": "Mackerson"
}
http://example.org/.../harvey.json input/ann.xml
Objects can be import from JSON, YAML, or RDF
36
<?xml version="1.0" encoding="UTF-8"?>
<rdf:RDF
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:sdo="https://schema.org/"
>
<rdf:Description rdf:about="https://peoples.r.us">
<sdo:givenName>Ann</sdo:givenName>
<rdf:type rdf:resource="https://schema.org/Person"/>
<sdo:familyName>Richardson</sdo:familyName>
<sdo:givenName>Elizabeth</sdo:givenName>
</rdf:Description>
</rdf:RDF>](https://image.slidesharecdn.com/linkmlyosemitepresentation-211018192042/85/LinkML-presentation-to-Yosemite-Group-36-320.jpg)