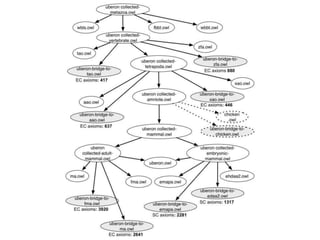
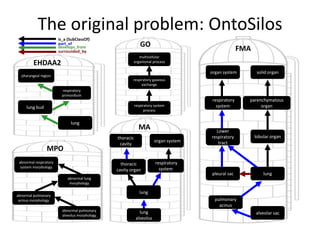
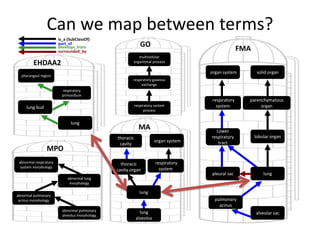
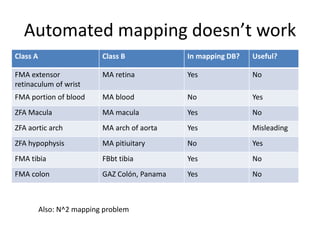
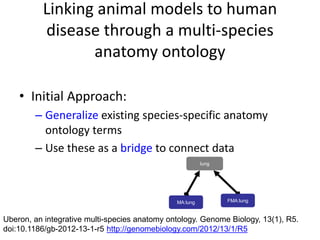
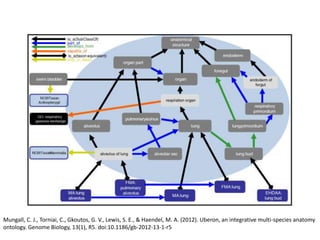
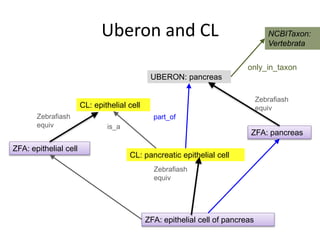
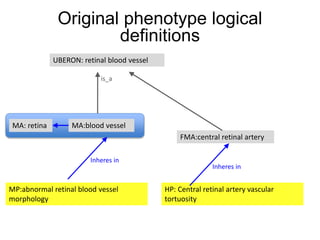
The document discusses Uberon, an integrative multi-species anatomy ontology. It describes Uberon's taxonomic scope covering metazoans with a focus on vertebrates. It outlines how Uberon is edited on GitHub and maintained with cross-references to other species-specific anatomy ontologies. It also discusses how phenotypes from the Phenotype and Human Phenotype Ontology are directly mapped to Uberon and species-specific anatomies, as well as considerations for which anatomy ontology a phenotype ontology should use.


![Editing Uberon
• GitHub repo layout predates ODK
– Some oddities!
• All content is in uberon_edit.obo
– [caveat: phenoscape and insecta extensions still to
be incorporated]
– Can be opened and edited in Protégé
• Anyone can make Pull Requests
– Travis-CI will check the PR
• Release cycle is semi-regular](https://image.slidesharecdn.com/uberon-potato-190408093330/85/Uberon-opening-up-to-community-contributions-12-320.jpg)