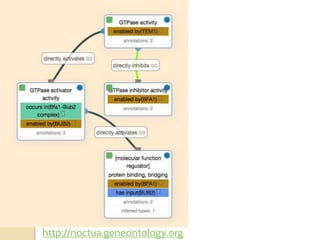
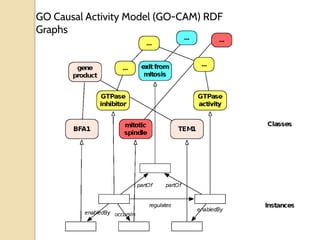
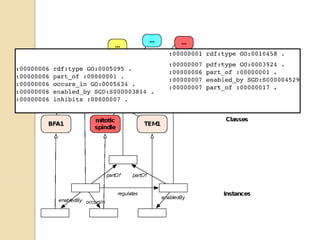
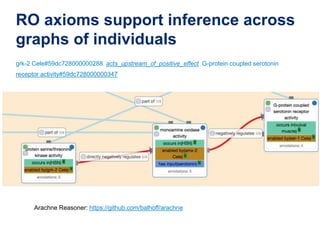
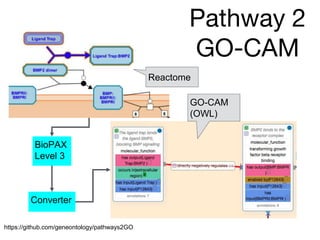
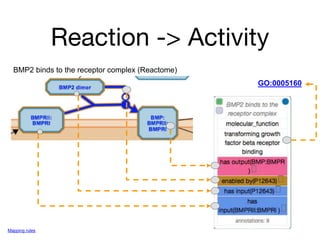
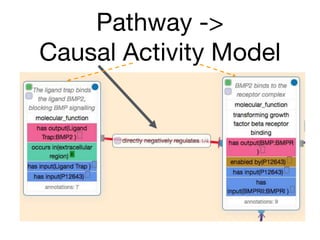
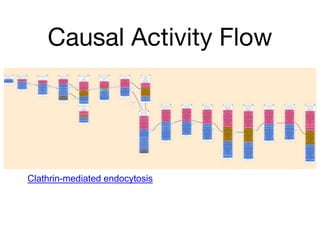
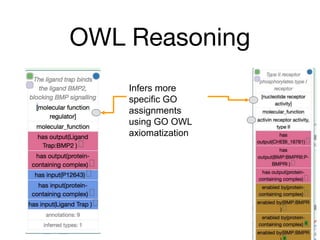
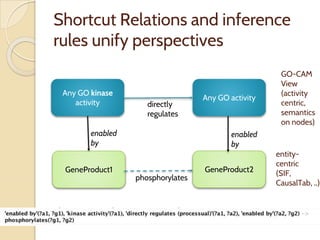
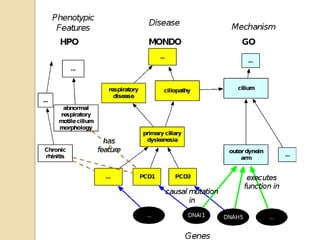
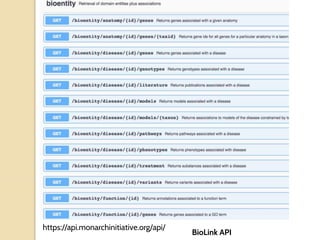
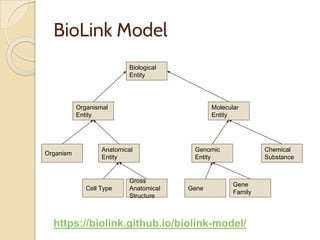
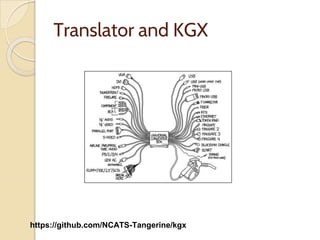
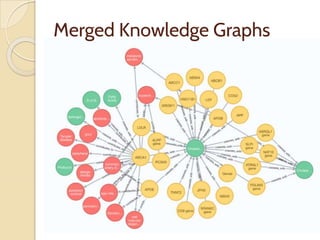
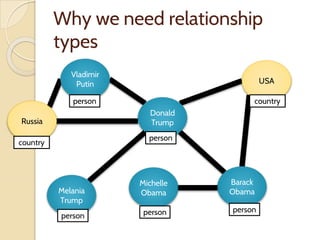
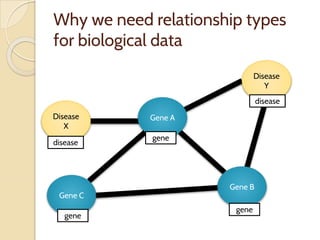
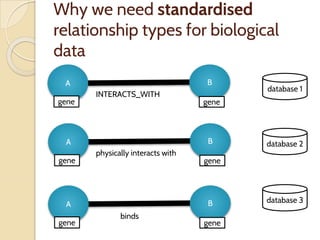
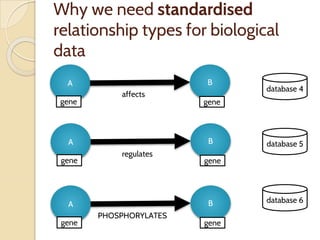
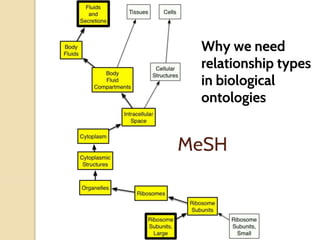
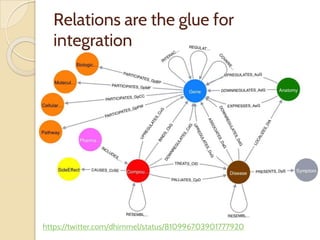
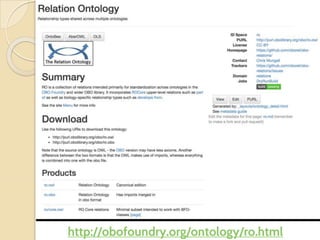
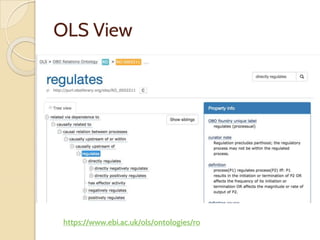
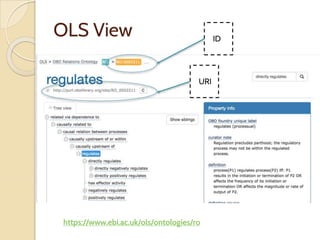
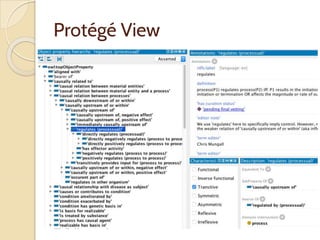
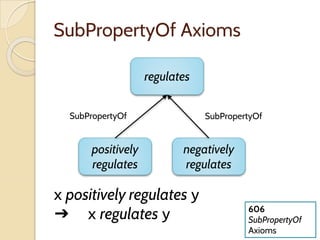
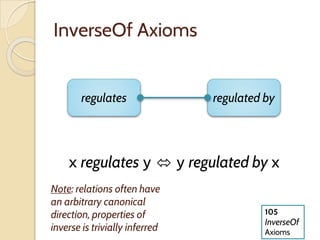
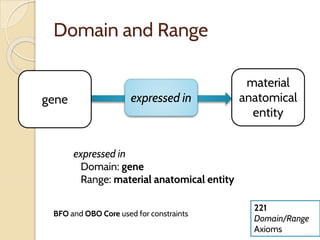
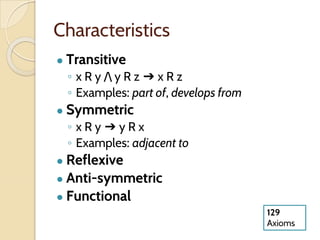
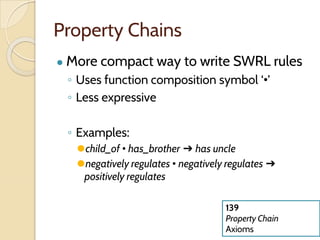
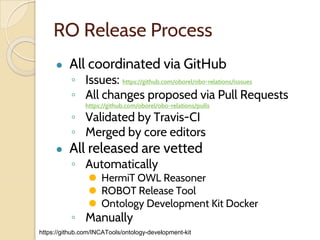
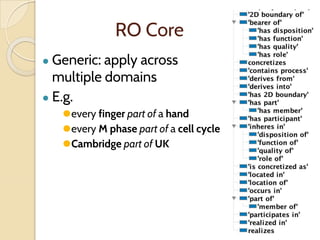
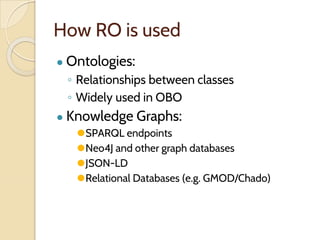
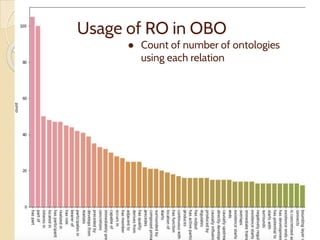
The document discusses the need for standardized relationship types in biological data and ontologies. It provides an overview of the Relation Ontology (RO), which defines over 450 standardized relationship types organized hierarchically. RO provides a foundation for integrating multiple knowledge graphs and represents relationships in ontologies, linked data, and knowledge bases. It enables logical reasoning and inference across graphs through properties like transitivity.










![GO’s initial attempts at
causality
GO:0086094
positive regulation of ryanodine-sensitive
calcium-release channel activity by
adrenergic receptor signaling pathway
involved in positive regulation of cardiac
muscle contraction
Mungall’s law[??*]: an inexpressive bio-database schema
will be abused to the maximum extent possible in order for
curators to express complexities of biology
[*] I have a feeling I’m not the first to express this
GO:0086023
adenylate cyclase-activating adrenergic receptor
signaling pathway involved in heart process
subClassOf](https://image.slidesharecdn.com/causal-reasoning-using-ro-2018-181015031000/85/Causal-reasoning-using-the-Relation-Ontology-32-320.jpg)