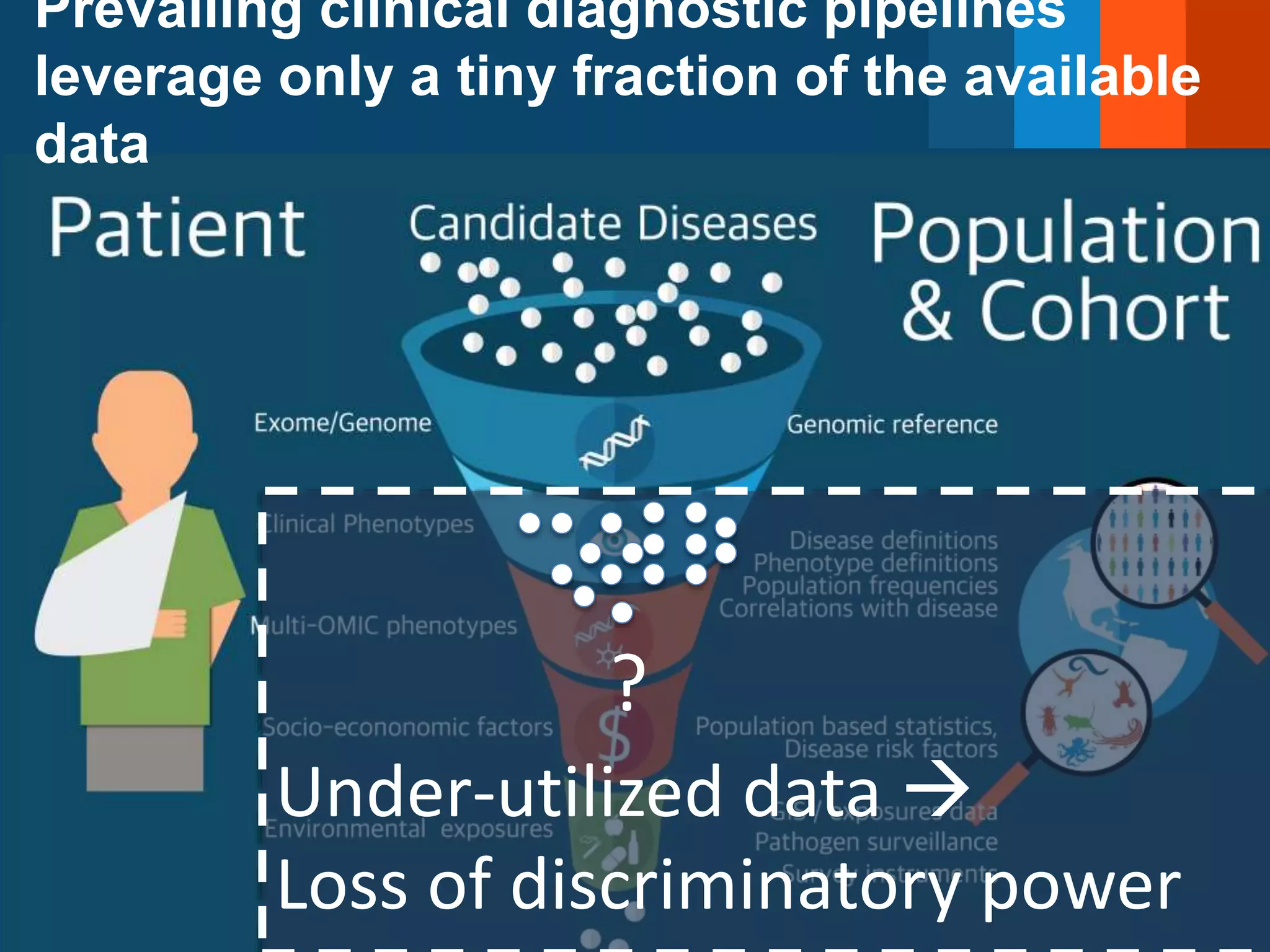
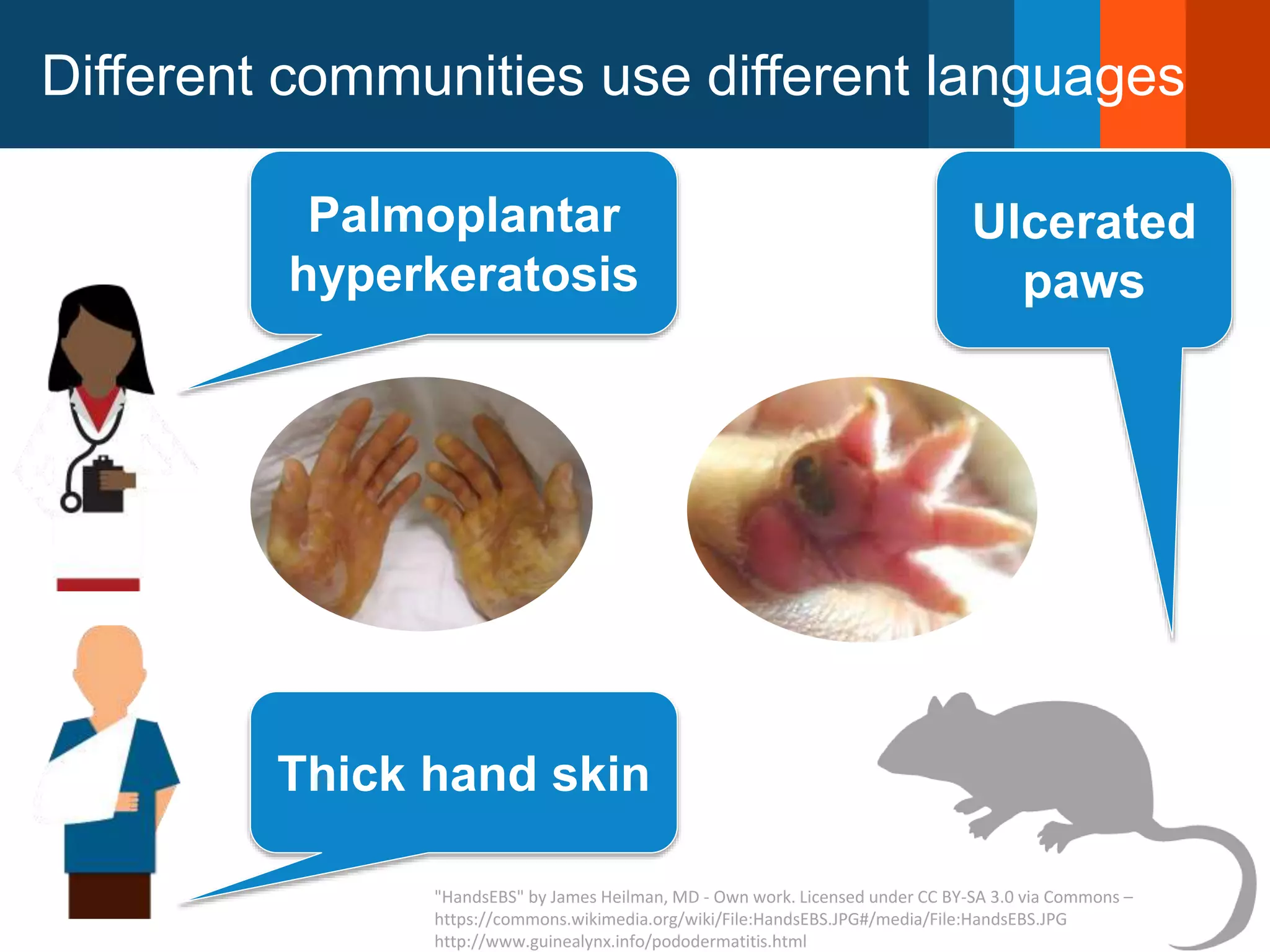
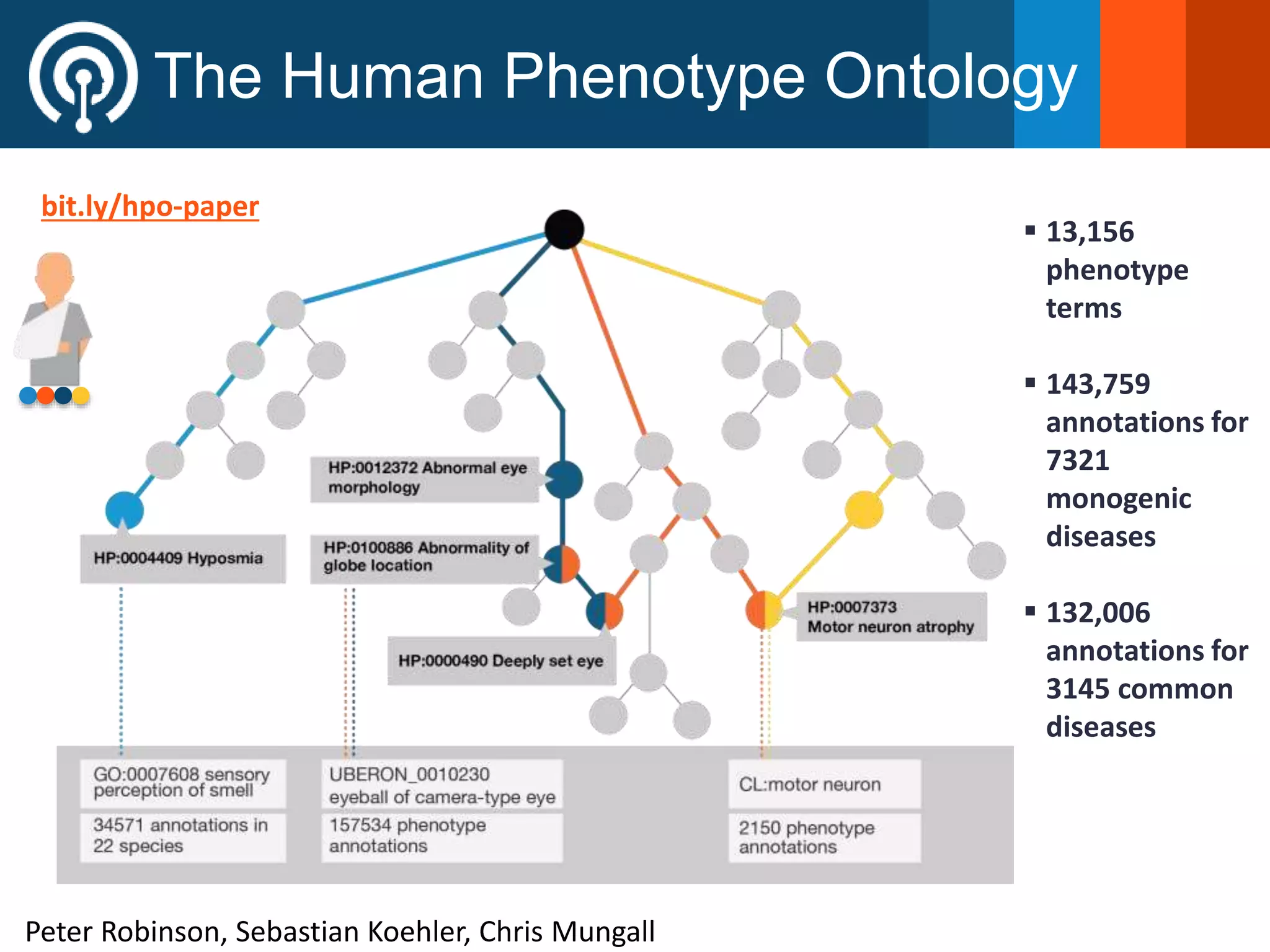
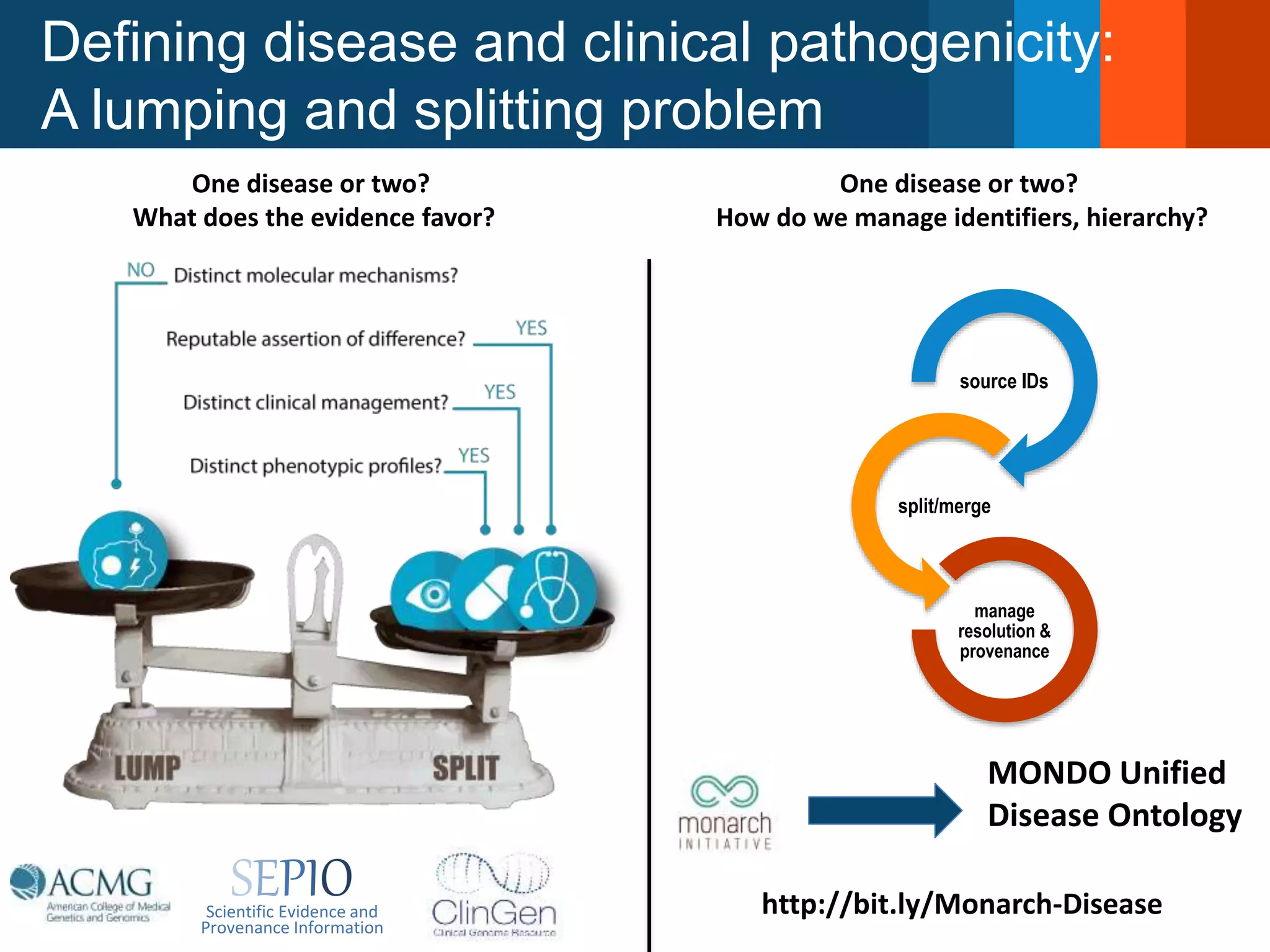
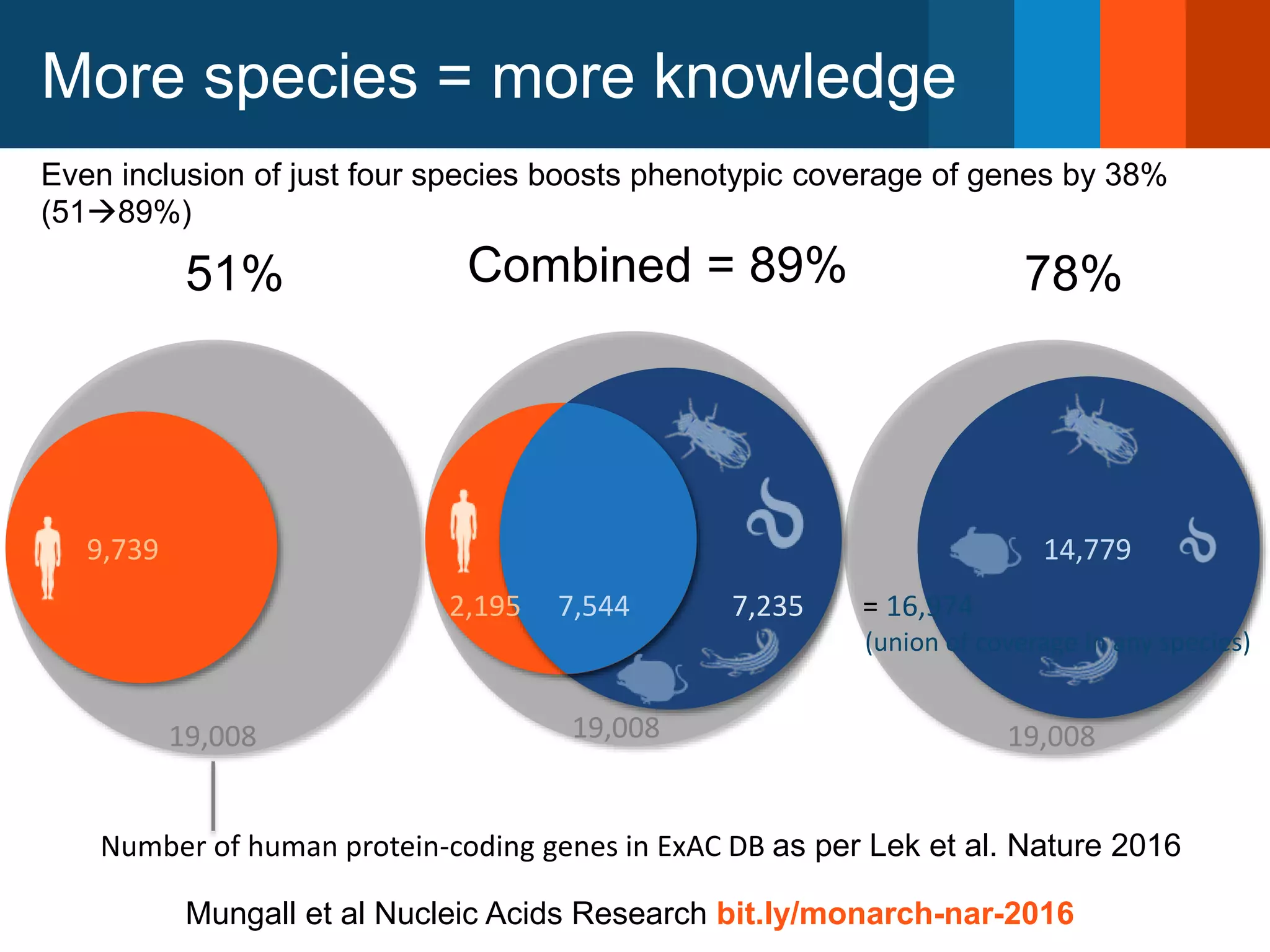
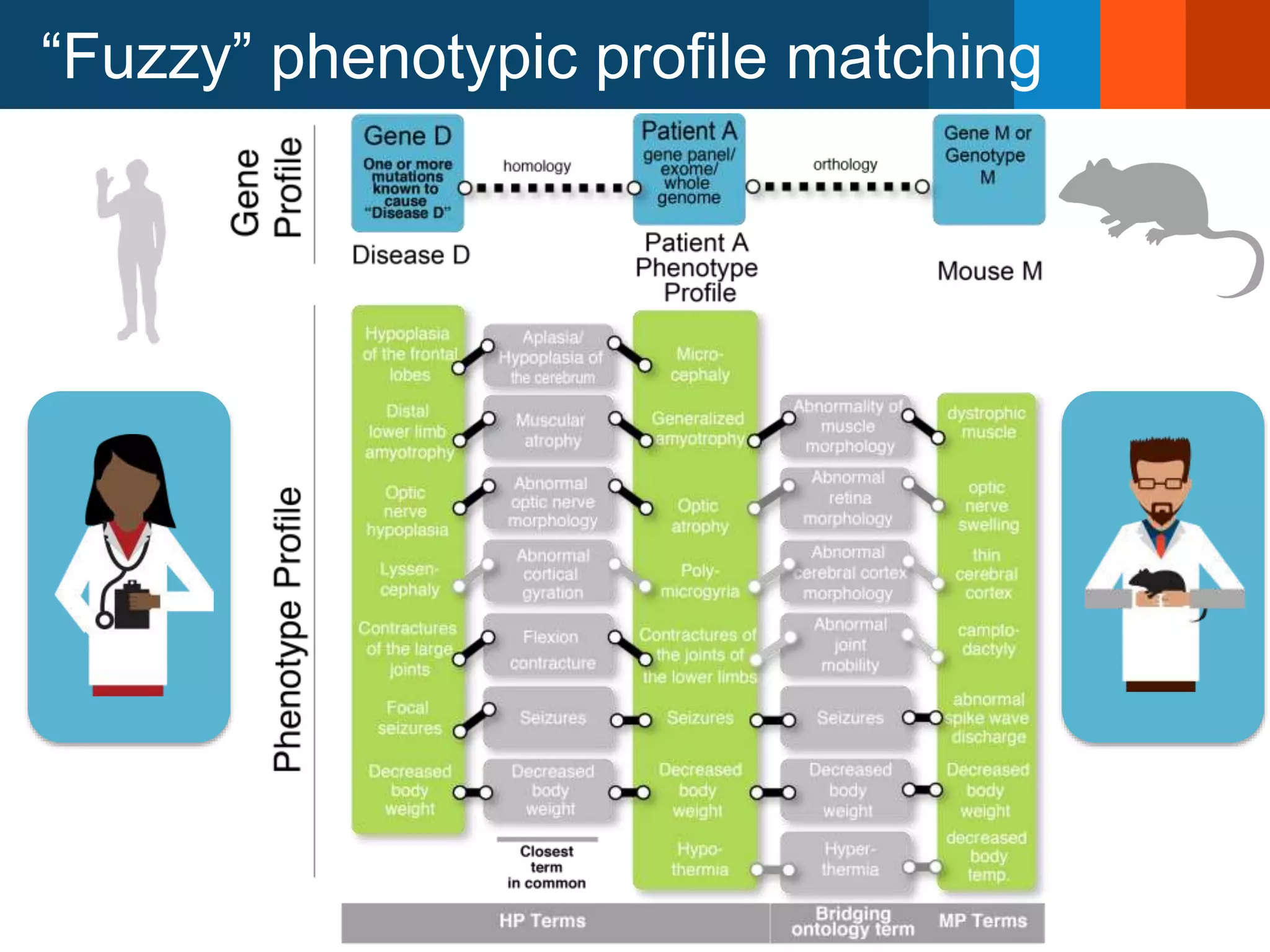
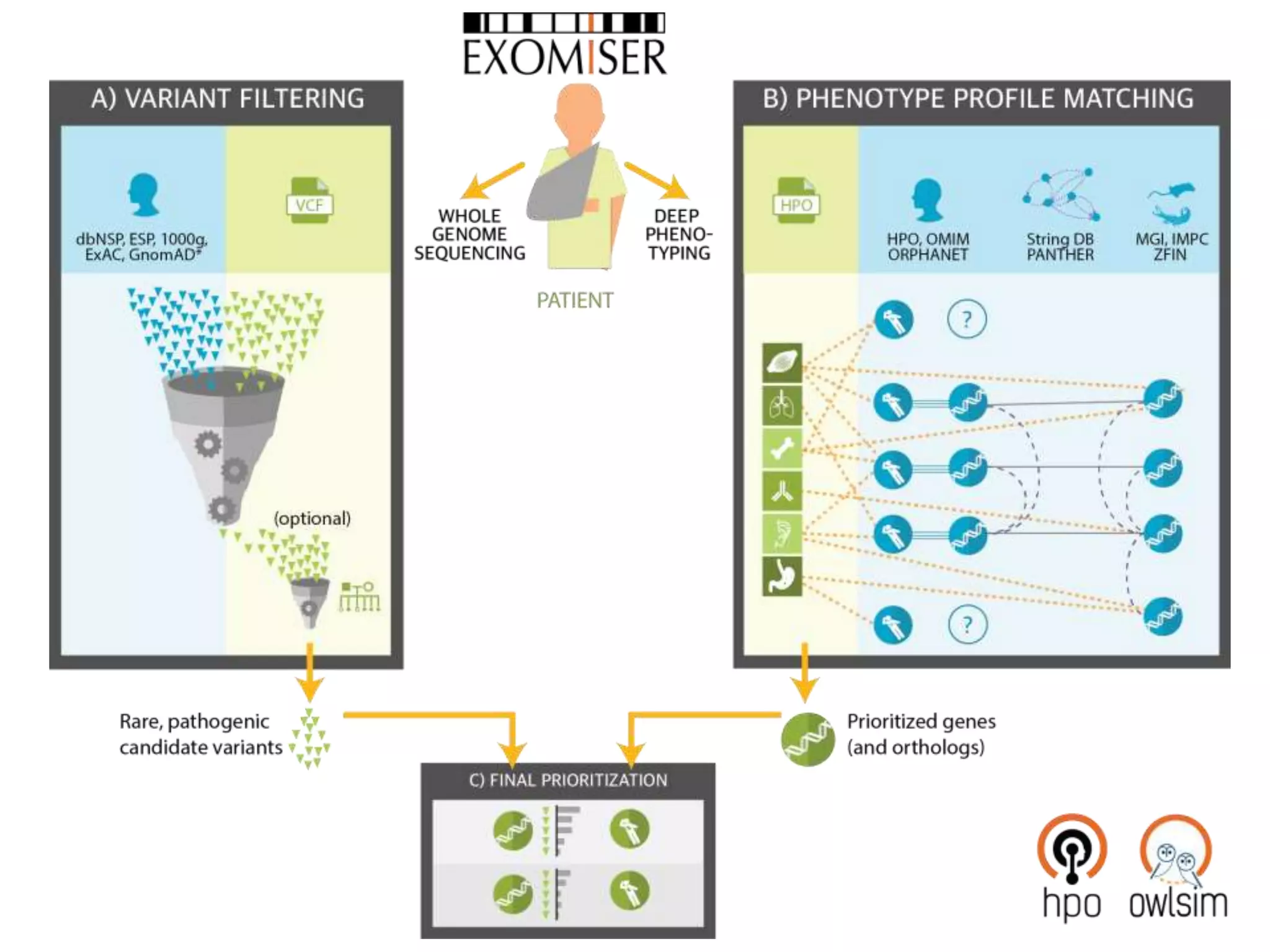
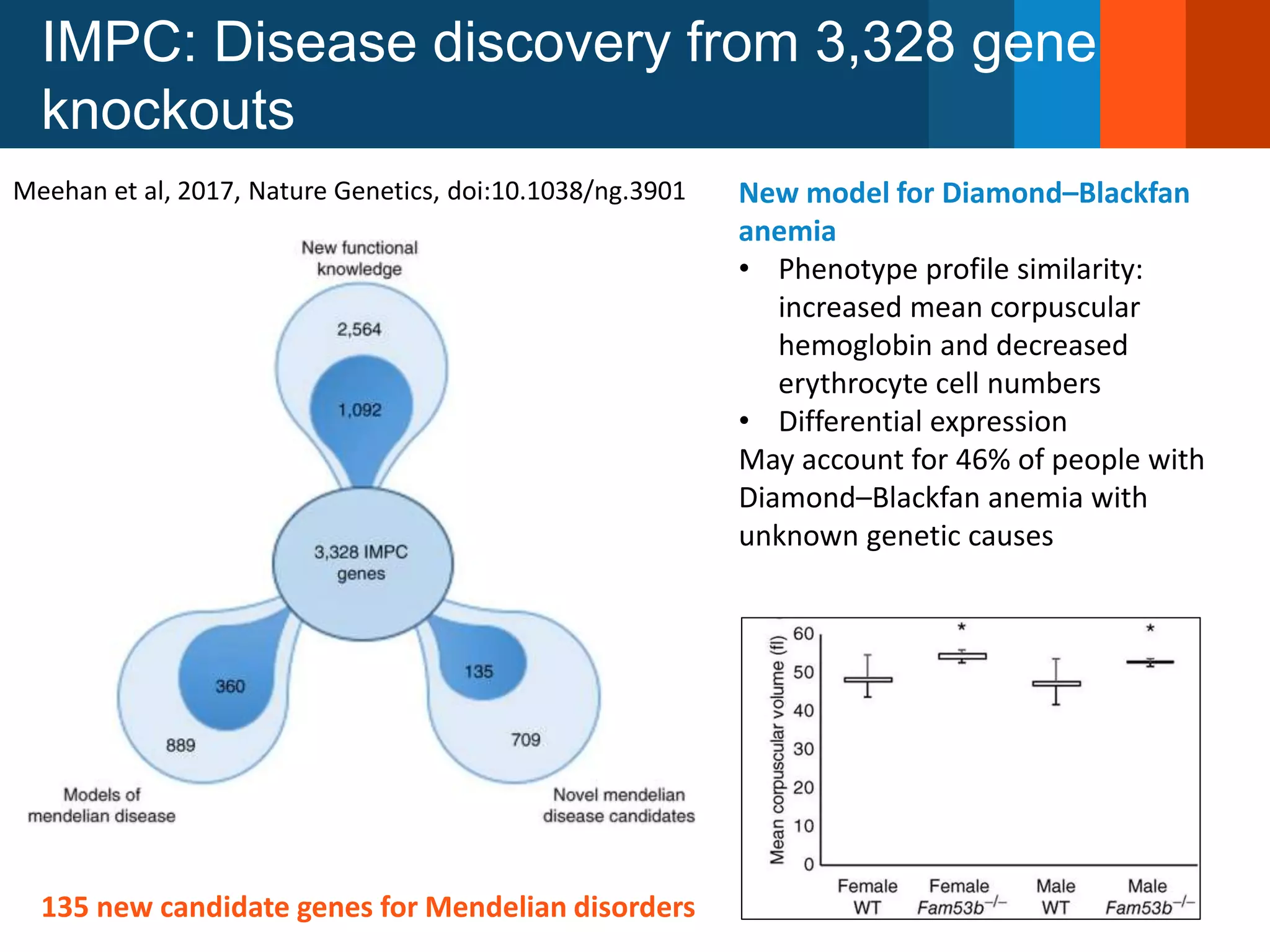
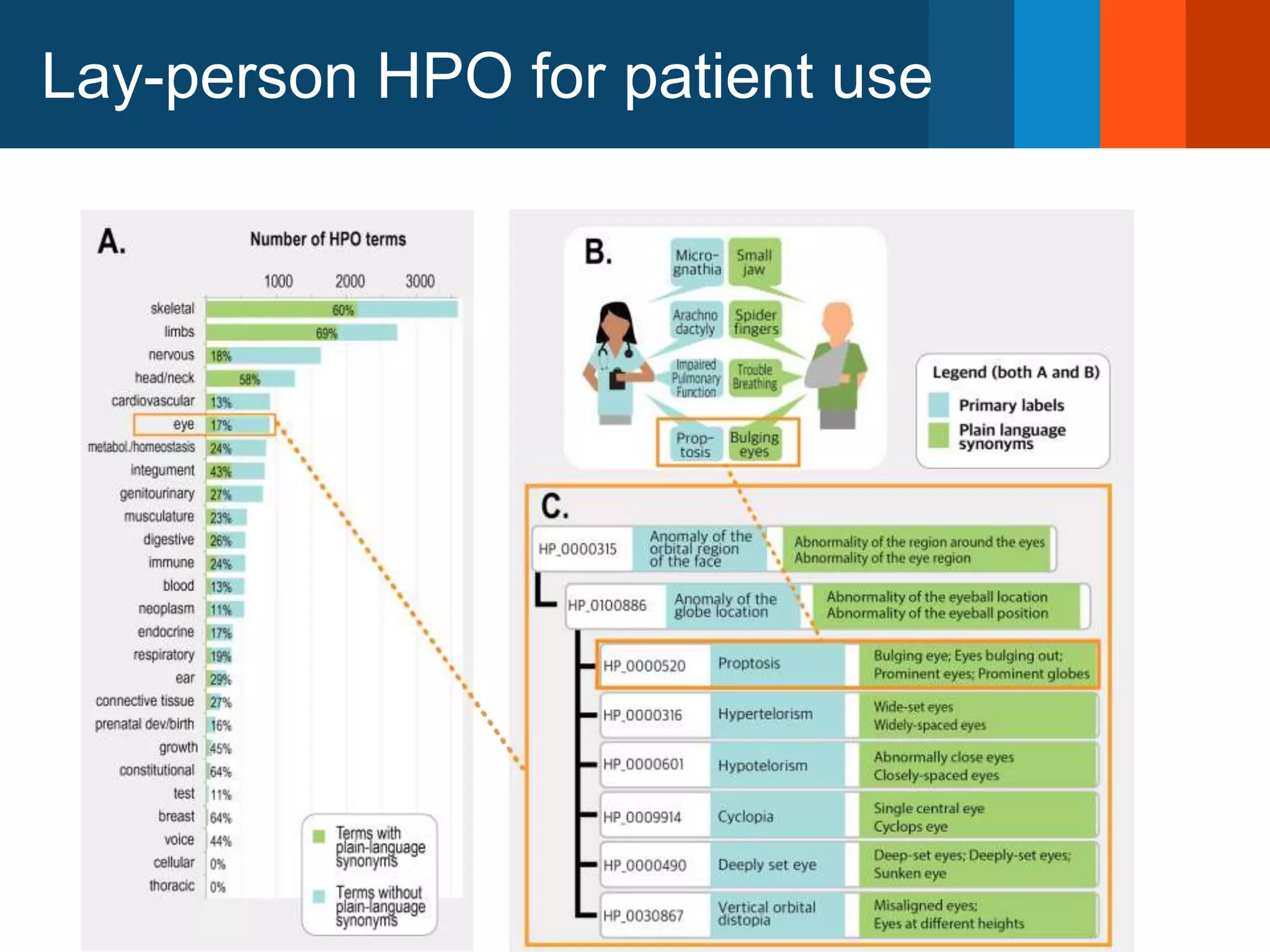
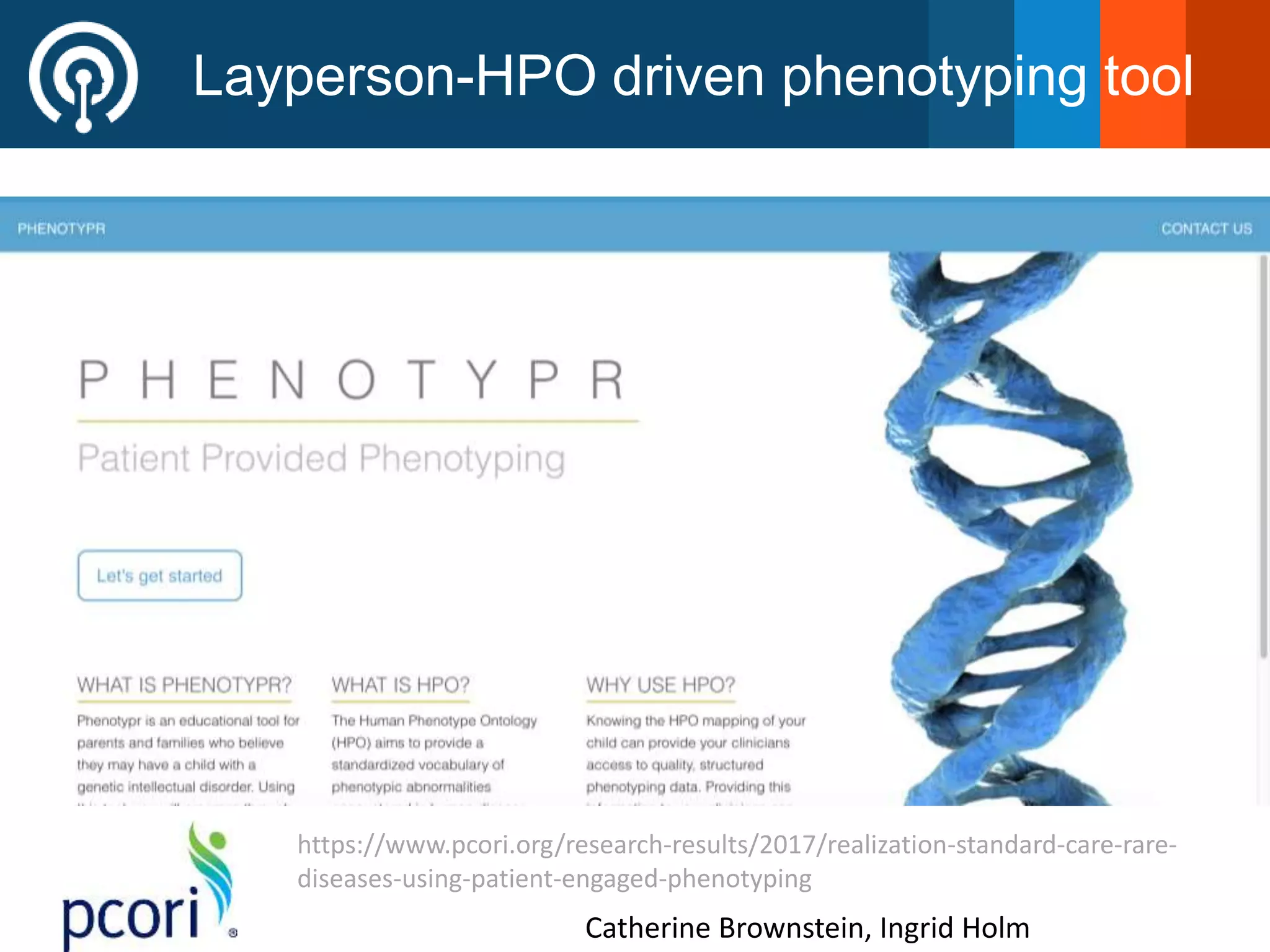
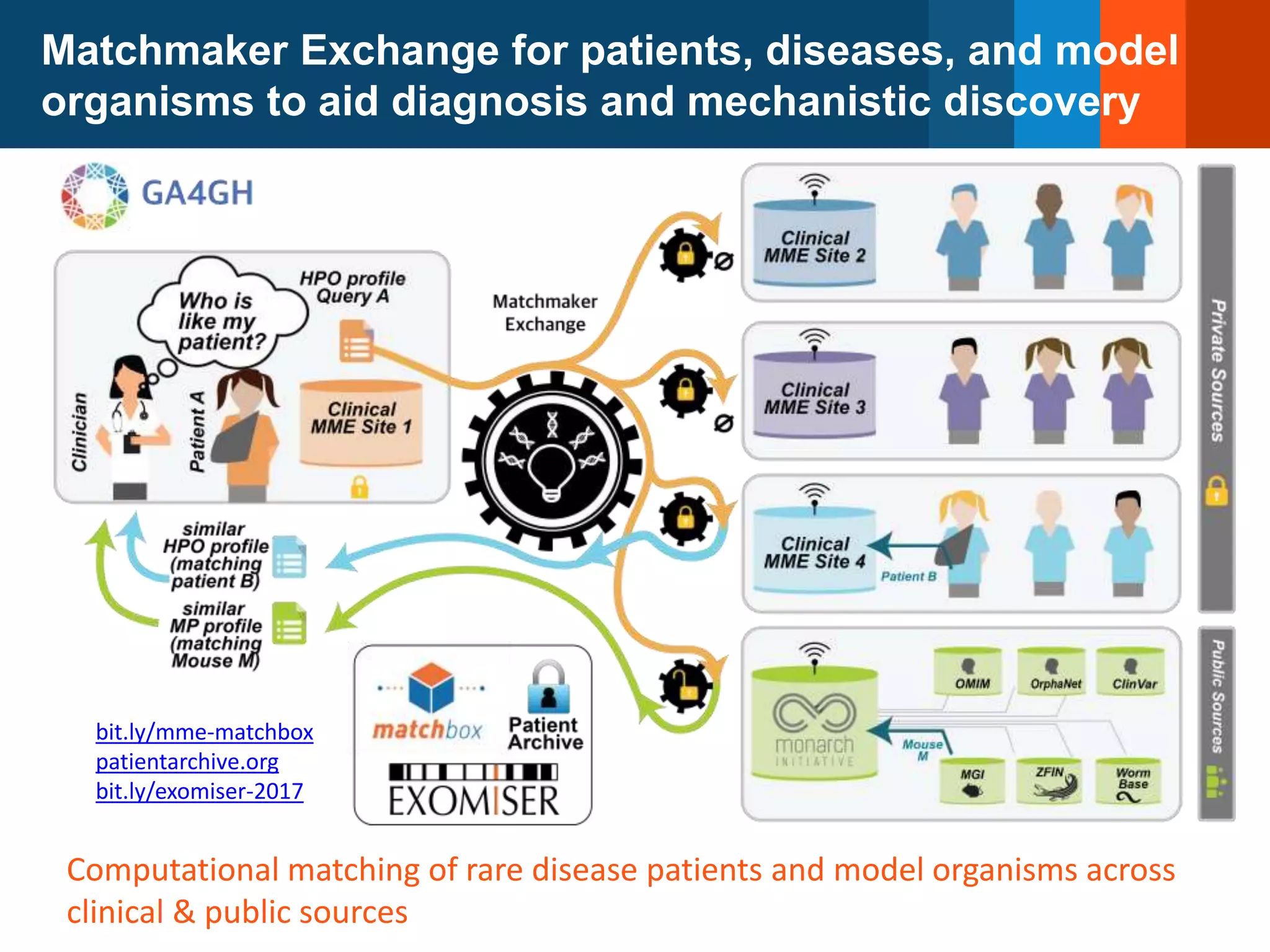
The Monarch Initiative aims to improve disease diagnostics and analysis by utilizing deep phenotyping data. It has developed ontologies like the Human Phenotype Ontology with over 13,000 phenotype terms to help machines understand human phenotypes. It uses "fuzzy" phenotypic profile matching across species to match patient data to known genetic disorders, as demonstrated by a case solved linking a patient's profile to a STIM1 variant. The Initiative is working to develop lay-friendly phenotyping tools and connect data sources through the Matchmaker Exchange to aid in diagnosis and research.