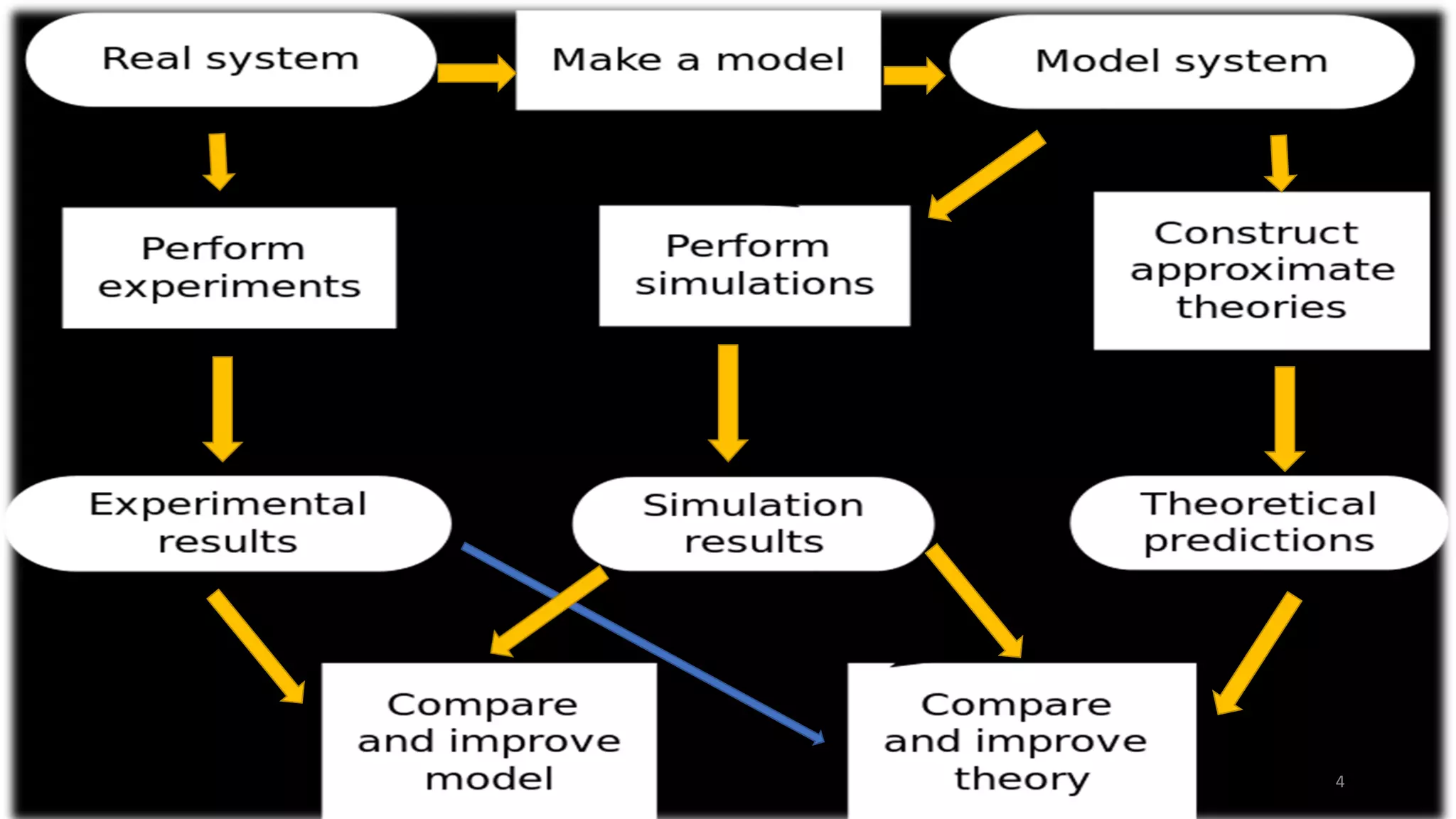
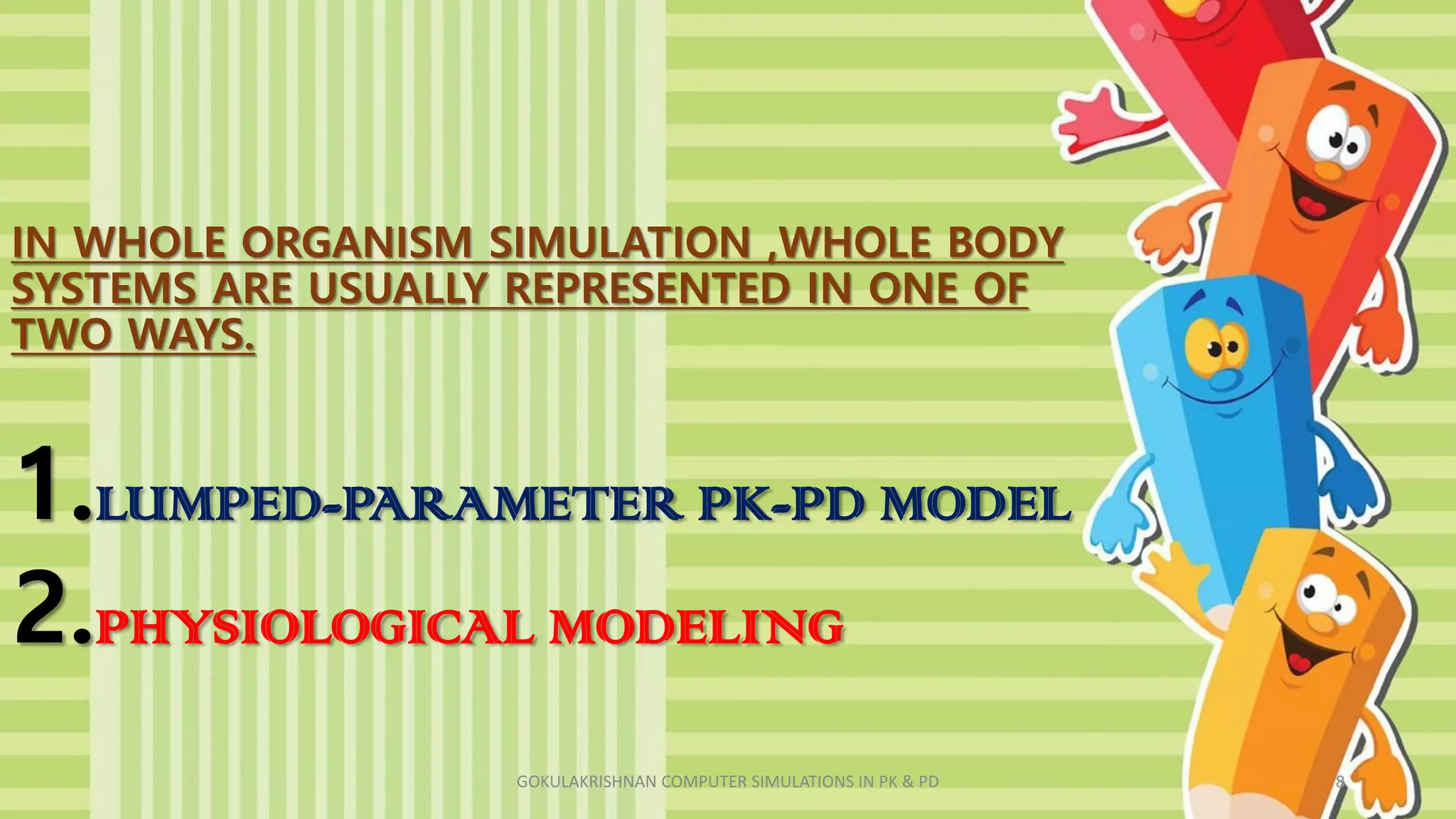
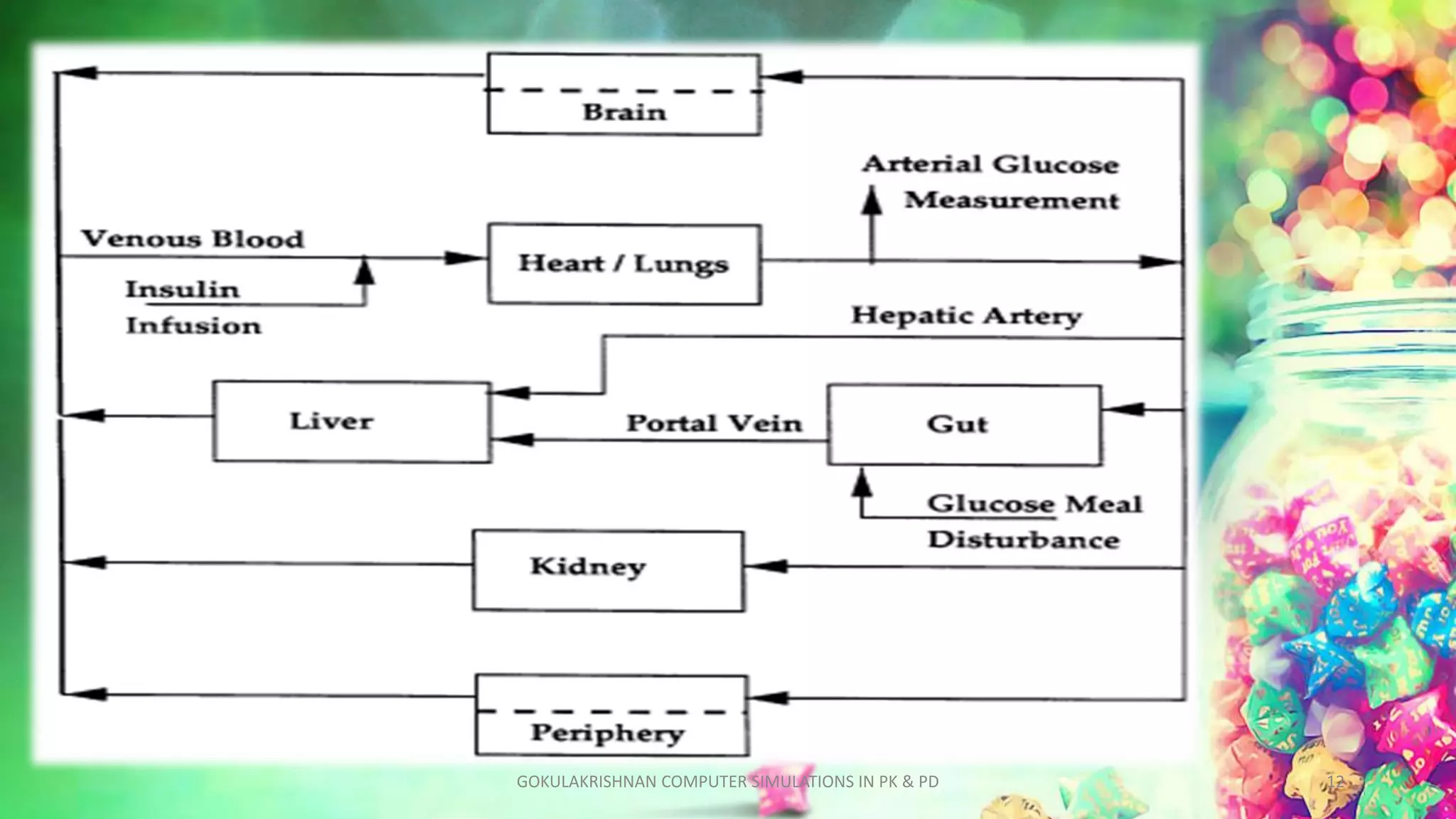
This document discusses computer simulations in pharmacokinetics and pharmacodynamics. It describes how whole organism, isolated tissue, and organ simulations work. For whole organism simulations, two approaches are used: lumped-parameter PK-PD modeling which uses differential equations to model the system over time, and physiological modeling which attempts to model interacting organs in detail. Isolated tissue and organ simulations are also discussed, focusing on models of the heart, liver, kidney and brain. The challenges of complexity and model selection are addressed.