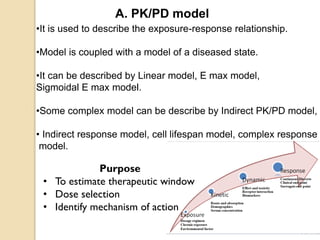
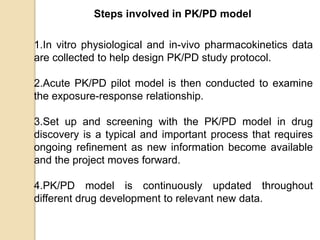
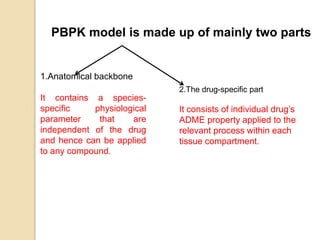
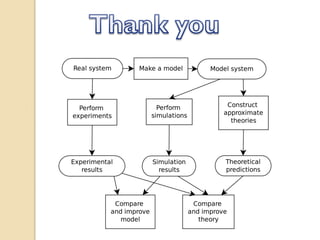
This document discusses computer simulation in pharmacokinetics and pharmacodynamics. It begins by defining computer simulation and explaining its importance in the biomedical field. It then describes the four levels of simulation: [1] simulation of the whole organism, [2] isolated tissues and organs, [3] the cell, and [4] proteins and genes. For whole organism simulation, it discusses physiologically-based pharmacokinetic (PBPK) models and pharmacokinetic-pharmacodynamic (PK/PD) models. The key steps in building PK/PD models are also outlined.