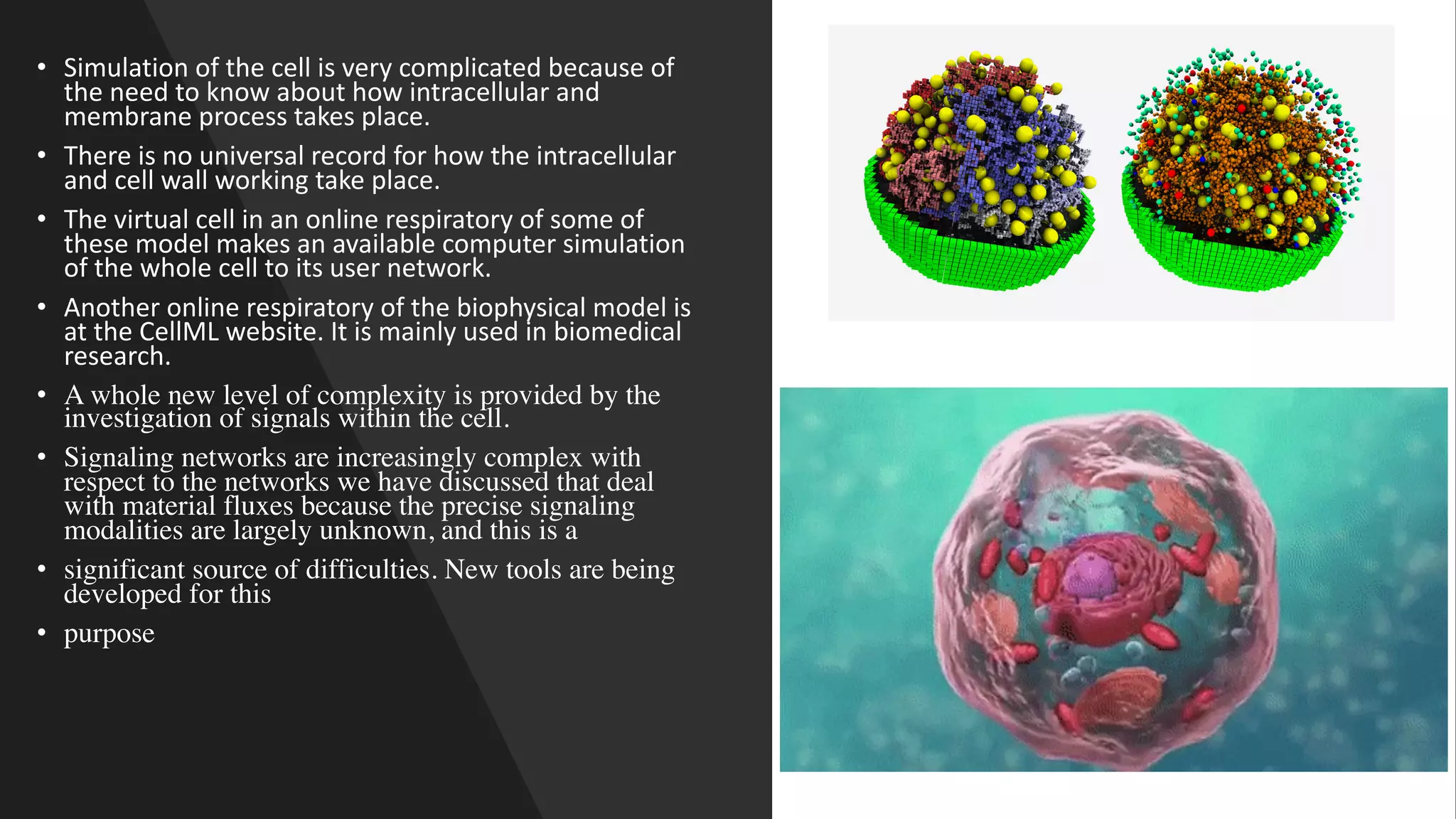
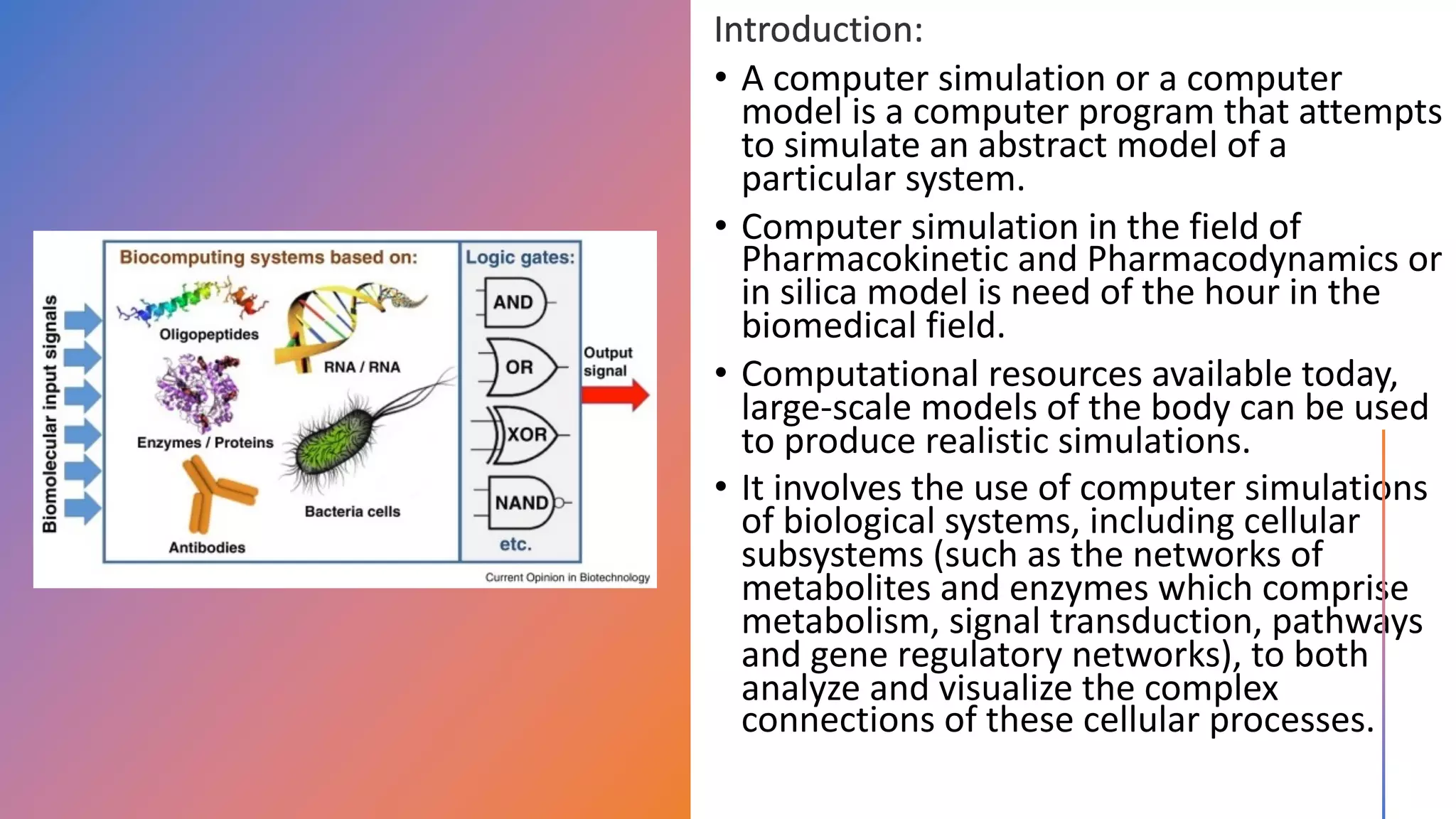
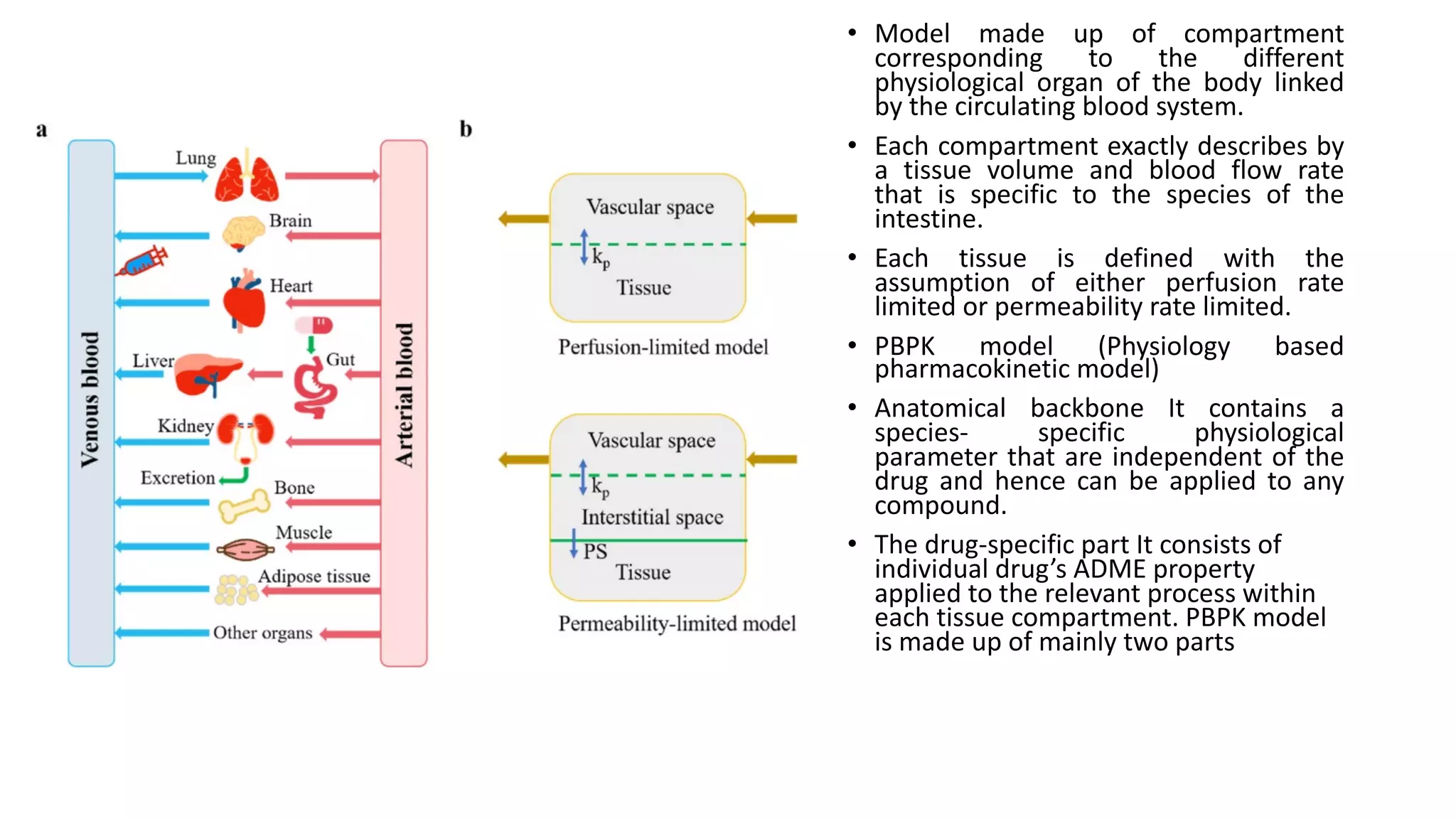
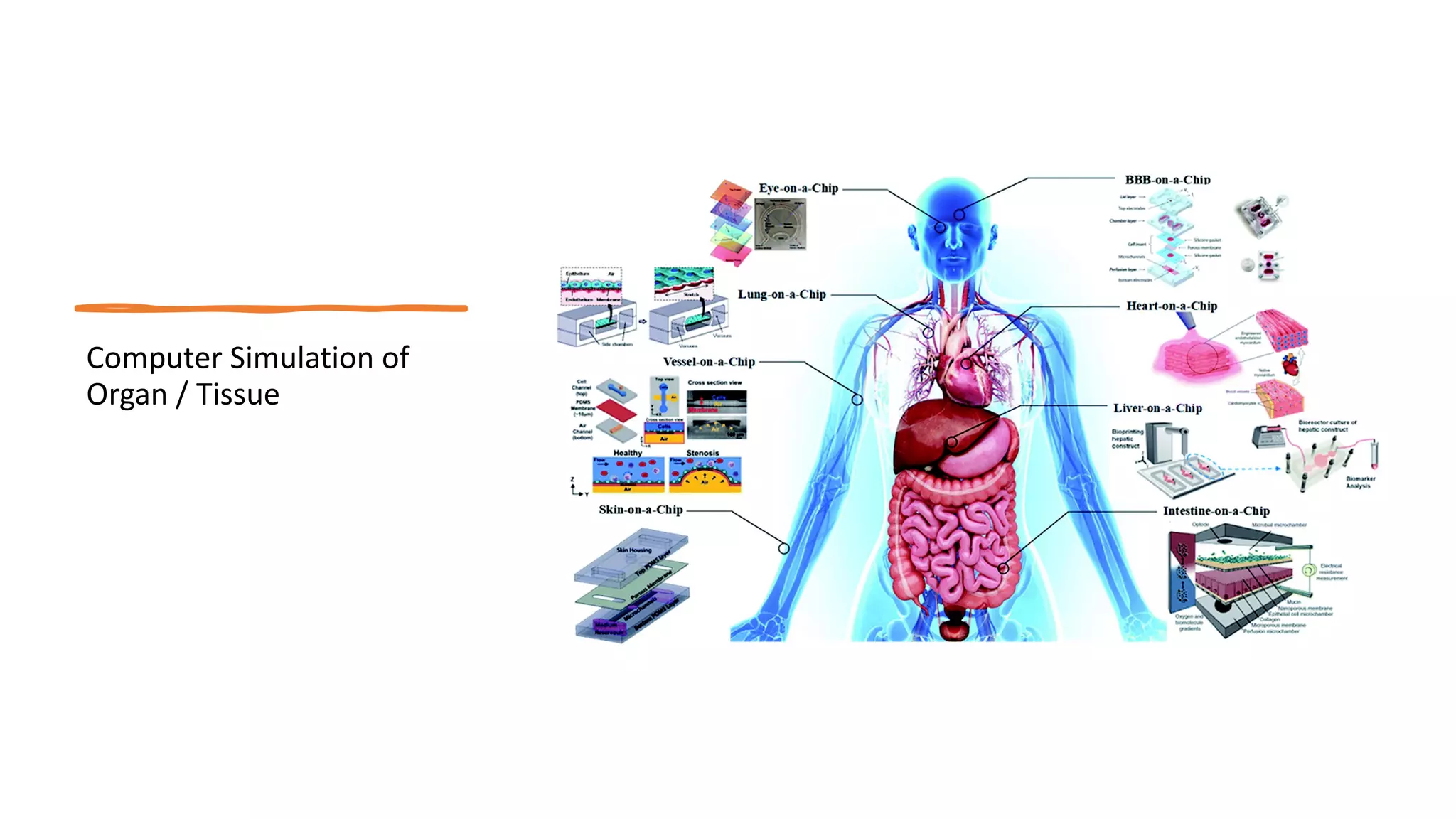
Computer simulations are increasingly used in pharmacokinetics and pharmacodynamics research. Simulations can model the whole organism, individual organs or tissues, cells, proteins, and genes. Whole organism simulations integrate models of organ systems to realistically simulate drug behavior in the body. Physiology-based pharmacokinetic models use anatomical and physiological parameters to model absorption, distribution, metabolism, and excretion of drugs. Organ and tissue simulations provide more detailed models of key organs like the liver and heart. Cell simulations model complex intracellular and membrane processes. Protein and gene simulations provide insights into molecular-level interactions. Computer models are valuable tools that integrate knowledge across biological scales to advance pharmaceutical sciences.





![• New project funded by the National Institute for General Medical Sciences at the NIH, the Center
for Modeling Integrated Metabolic Systems (MIMS) [41], has as its mission the development and
integration of in vivo, organ-specific mathematical models that can successfully predict behaviors
for a range of parameters,including rest and exercise and various pathophysiological conditions.
• The Microcirculation Physiome [42] and the Cardiome [43] are other multicenter projects focused
on particular aspects of the Physiome undertaking.
• It seems widely accepted that the development of integrated computational representations of
biological systems has to borrow from many fi elds, if nothing else because of the
multidisciplinary complexity that some of these endeavor simply.](https://image.slidesharecdn.com/wholeorgancadds-220722083502-b1fd1095/75/COMPUTER-SIMULATIONS-IN-PHARMACOKINETICS-AND-PHARMACODYNAMICS-11-2048.jpg)