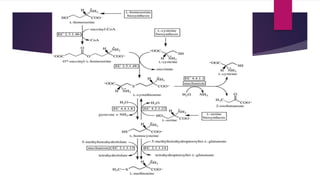
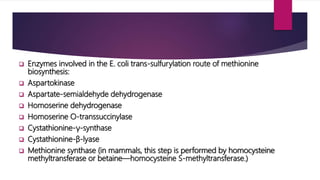
L-methionine is an essential amino acid involved in protein biosynthesis and various cellular functions. Bacteria, plants, and other organisms have different biosynthetic pathways for methionine, utilizing either transsulfuration or sulfhydrylation methods. Enzymes such as aspartokinase and methionine synthase play critical roles in the trans-sulfurylation route, particularly in Escherichia coli.