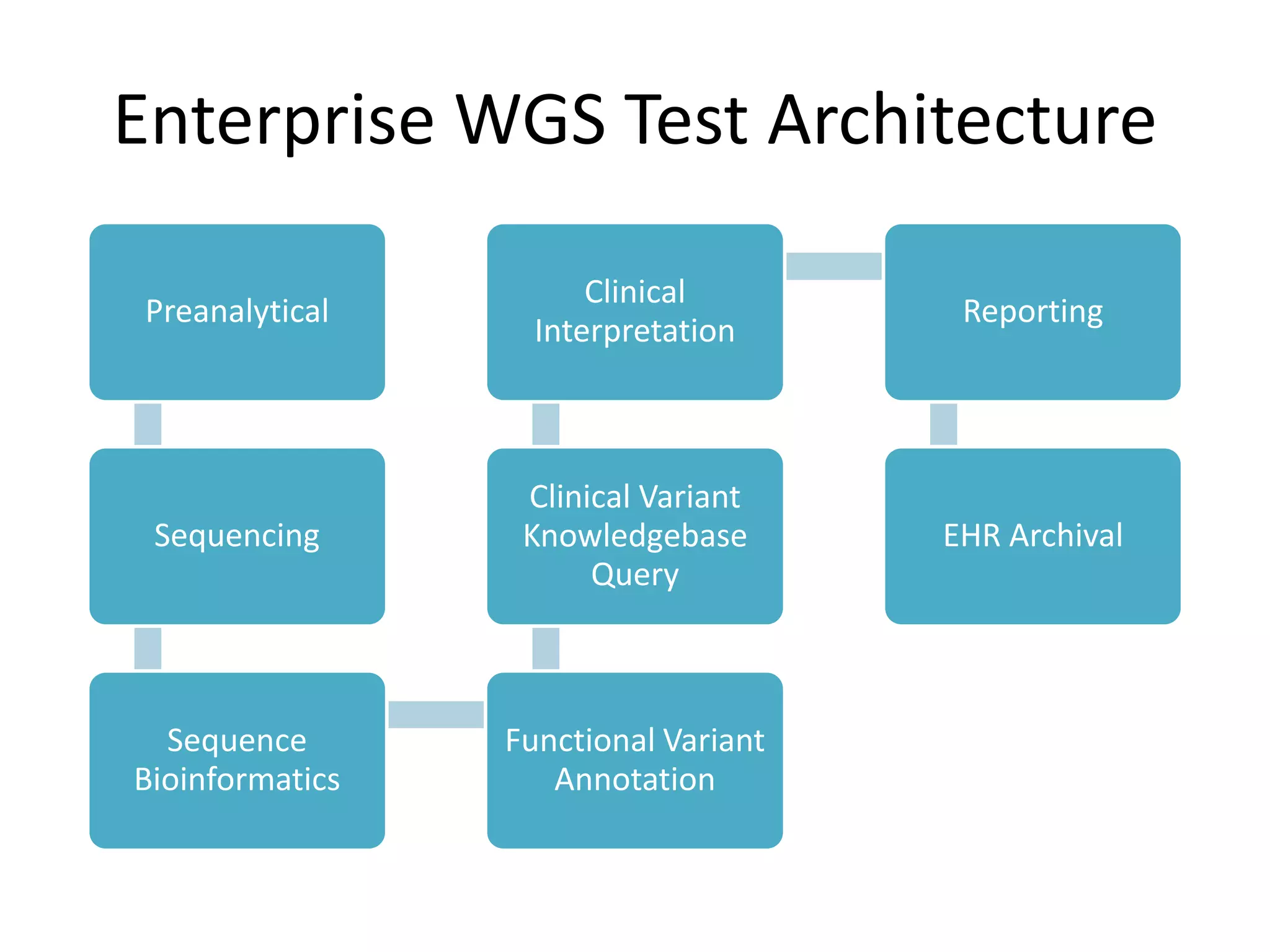
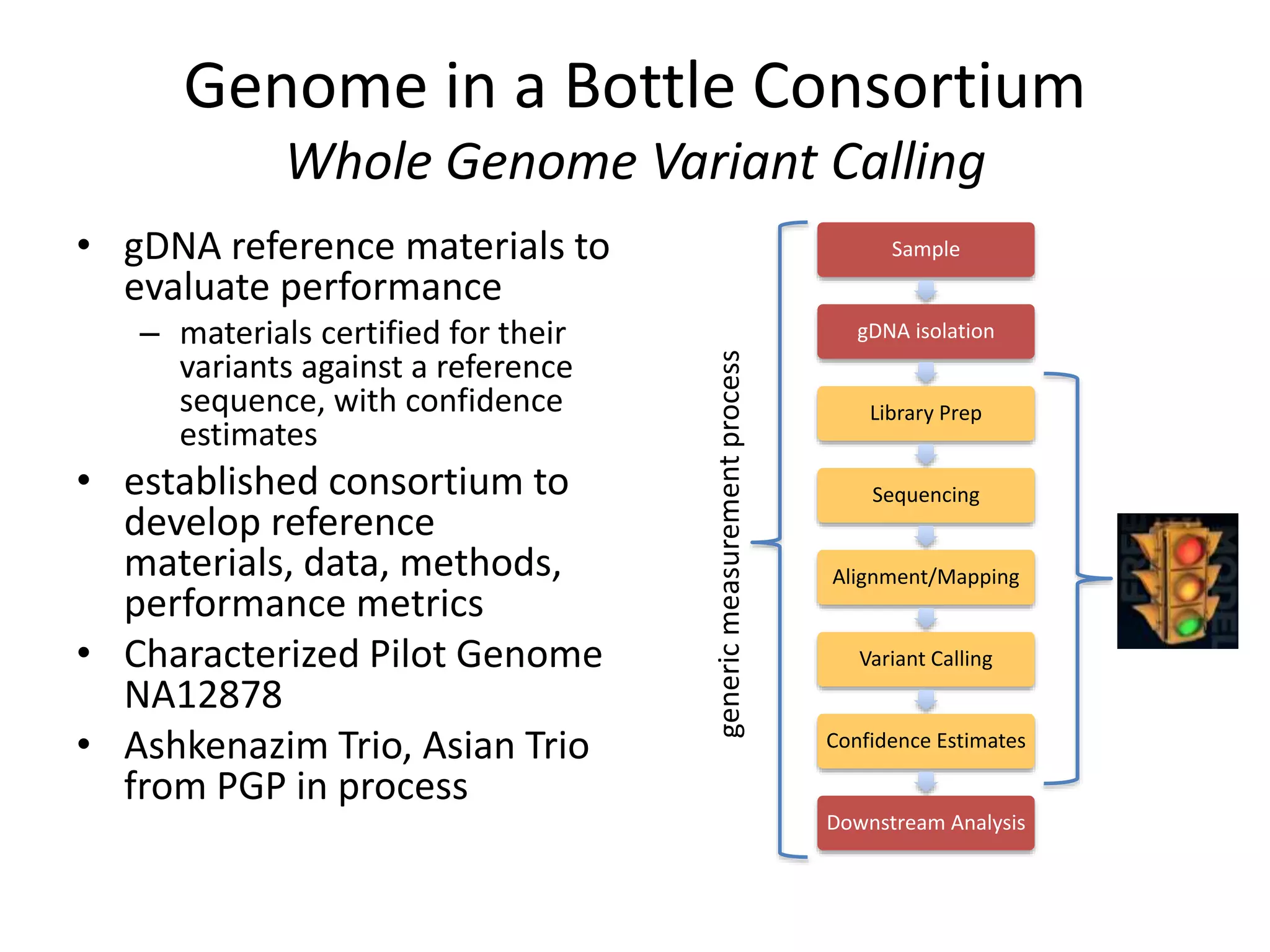
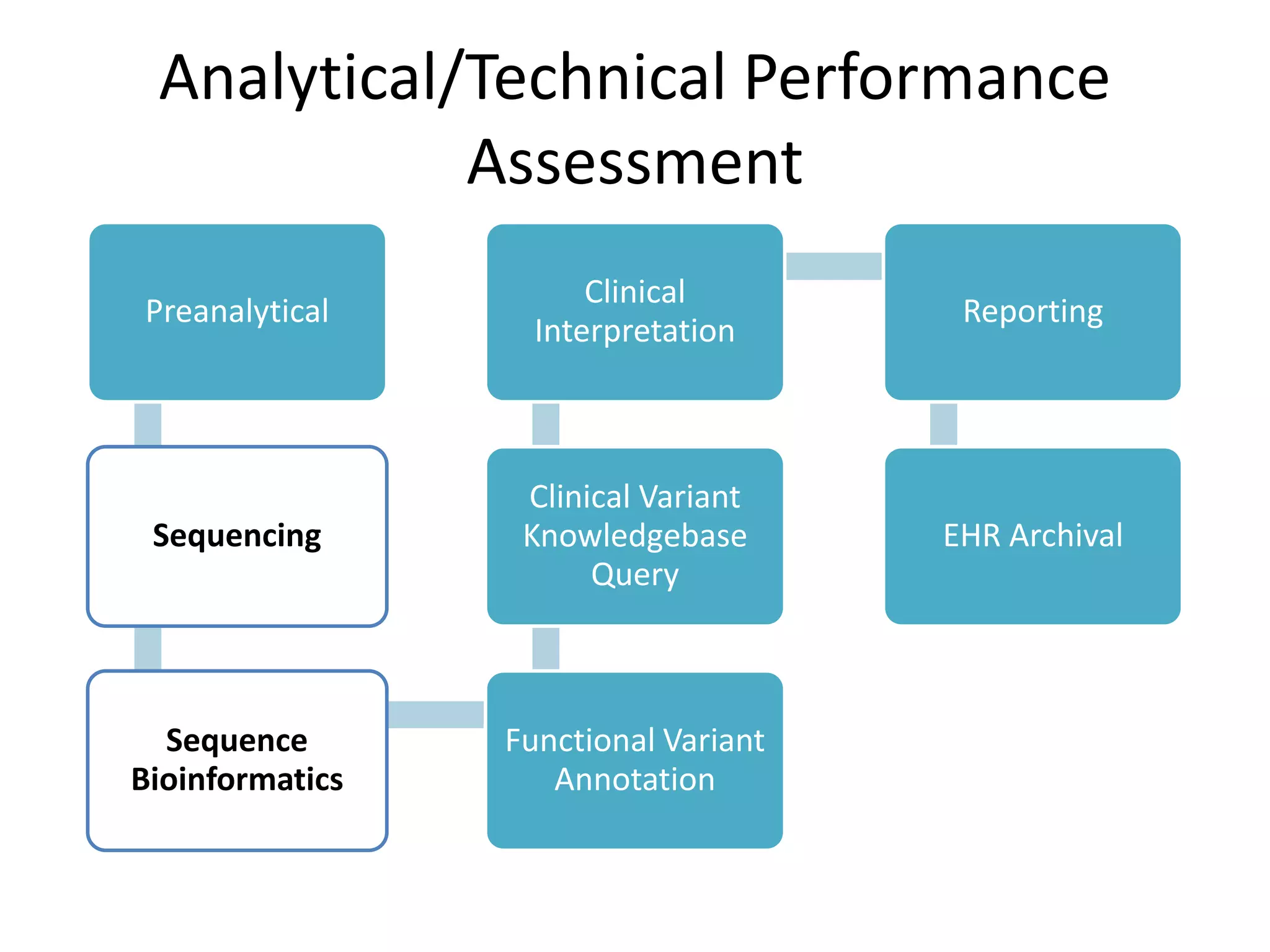
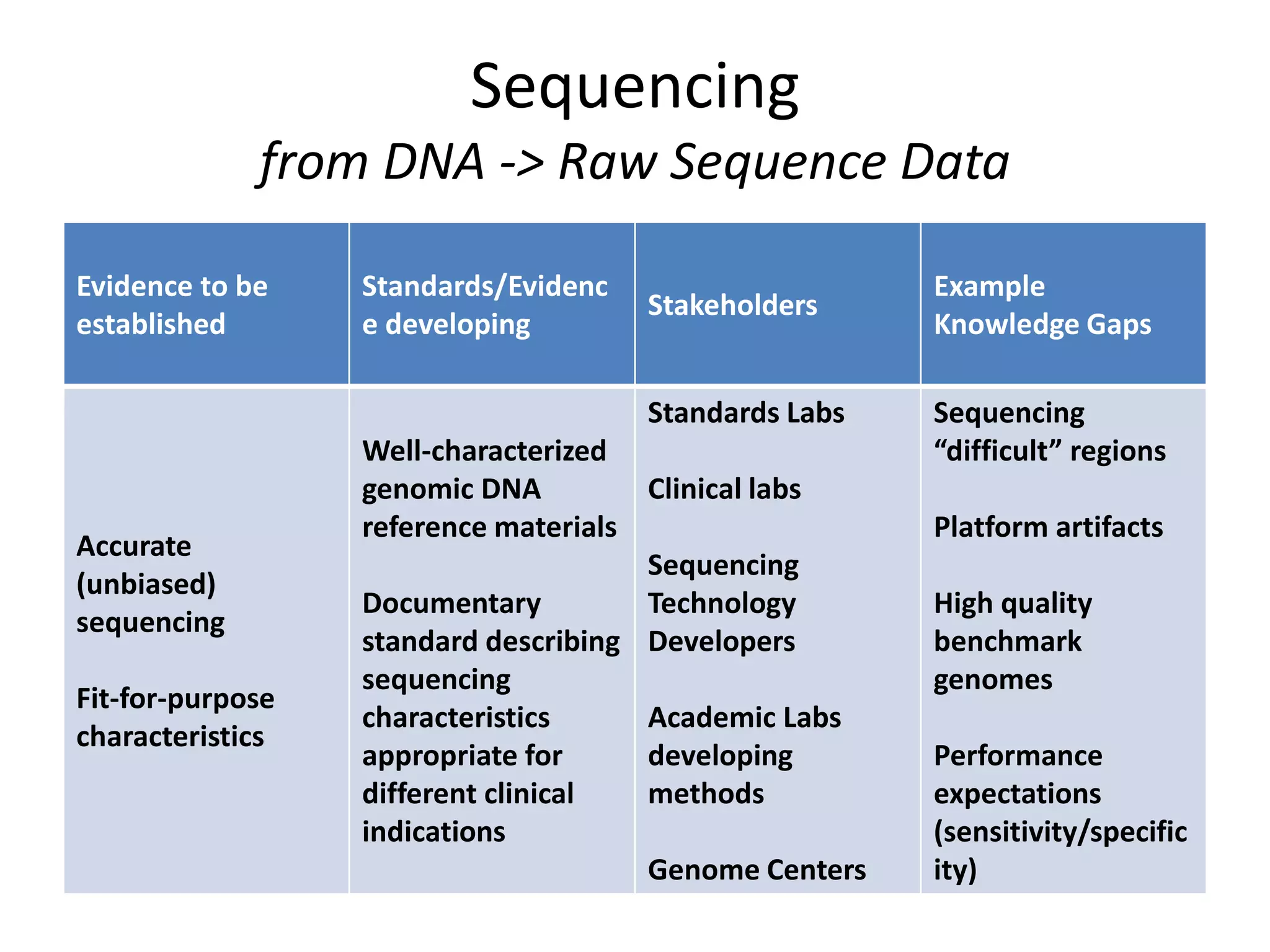
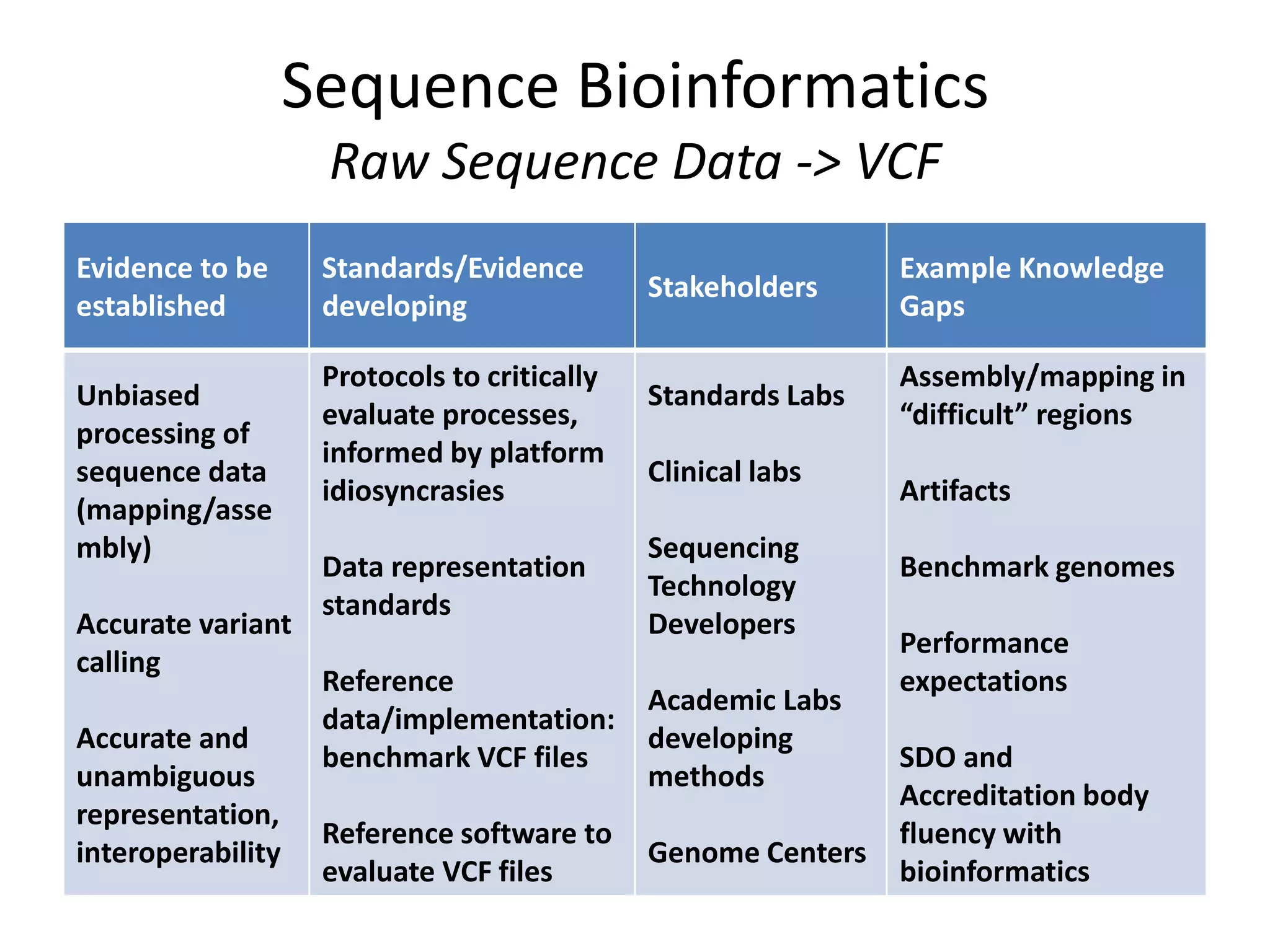
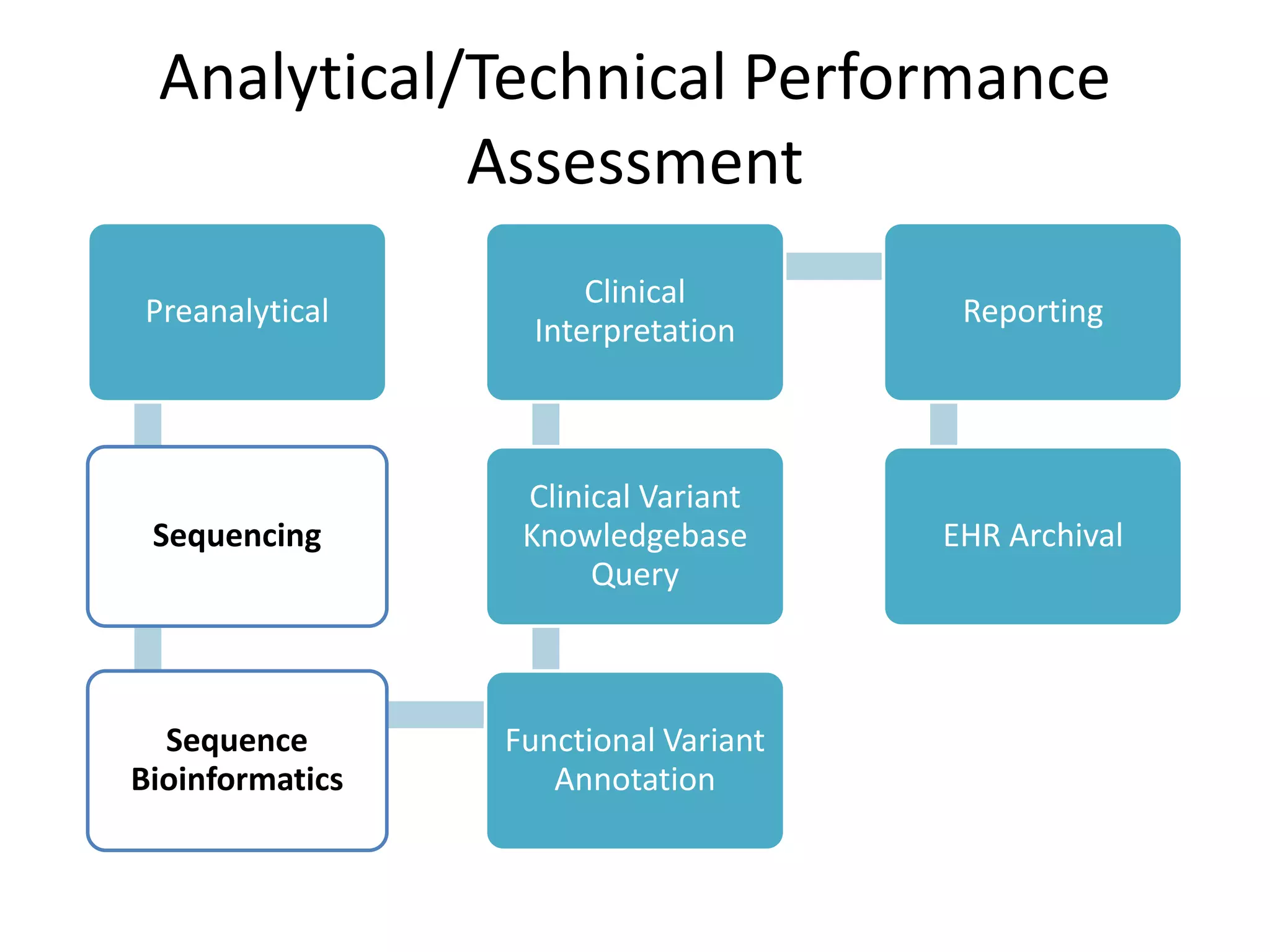
This document discusses standards for characterizing whole genome sequencing. It describes the Genome in a Bottle Consortium which aims to develop reference materials, data, methods and performance metrics for whole genome variant calling. The consortium has characterized the genome of sample NA12878 and is working on Ashkenazim and Asian trios. The document outlines the need for standards and evidence regarding accurate and unbiased sequencing, sequence bioinformatics processing, and functional and clinical variant annotation in order to assess analytical and technical performance of whole genome sequencing for clinical use.