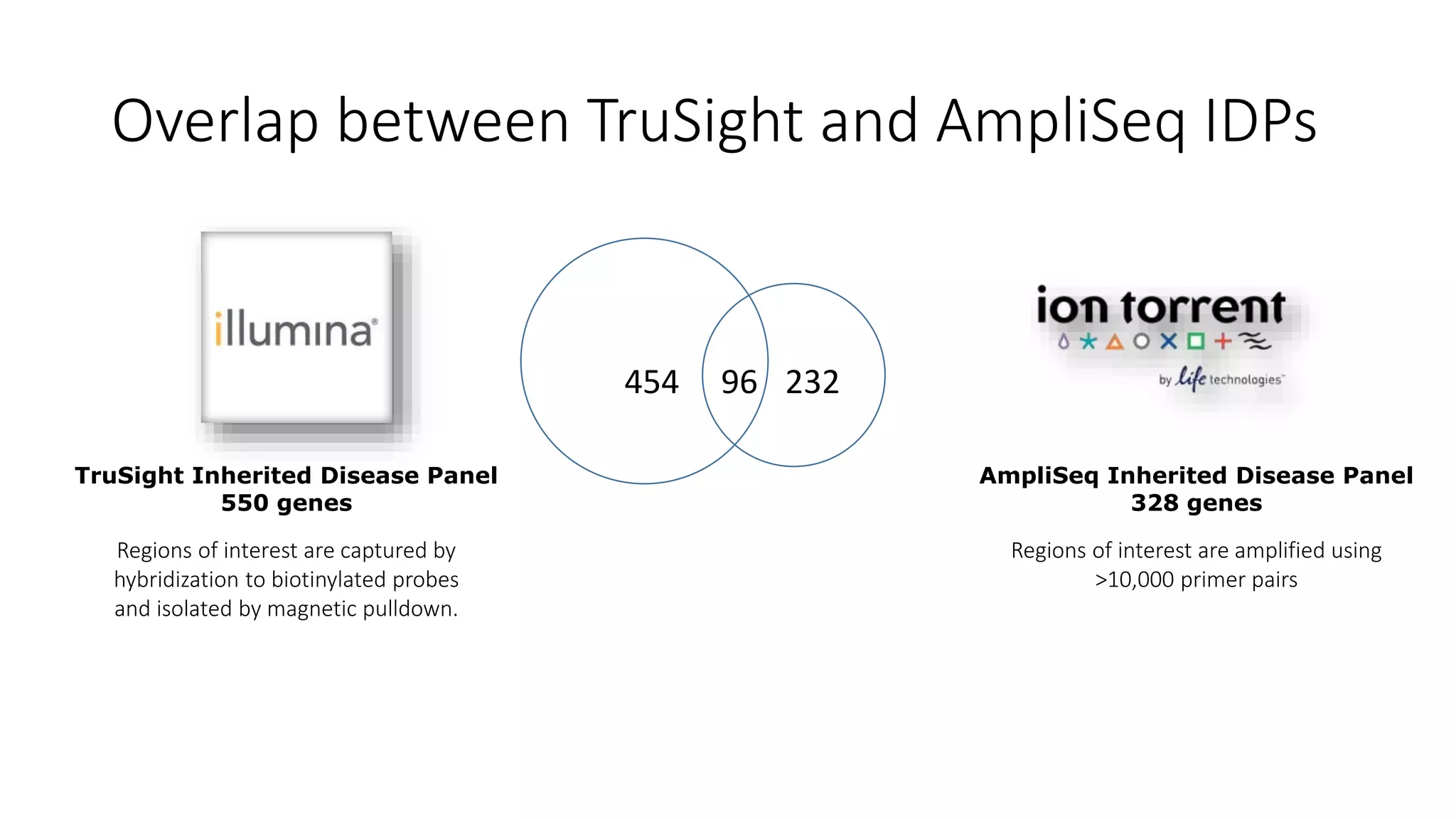
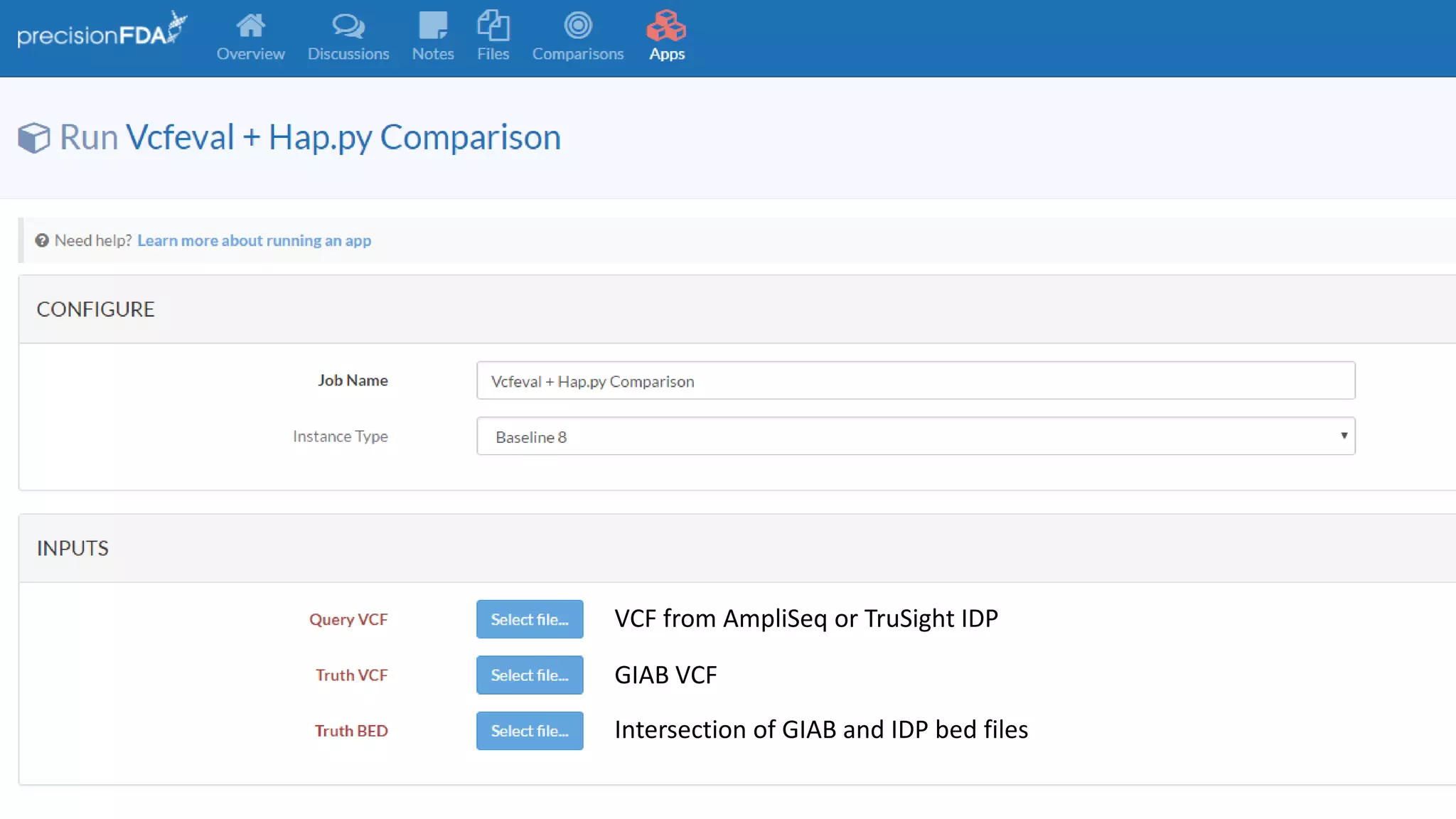
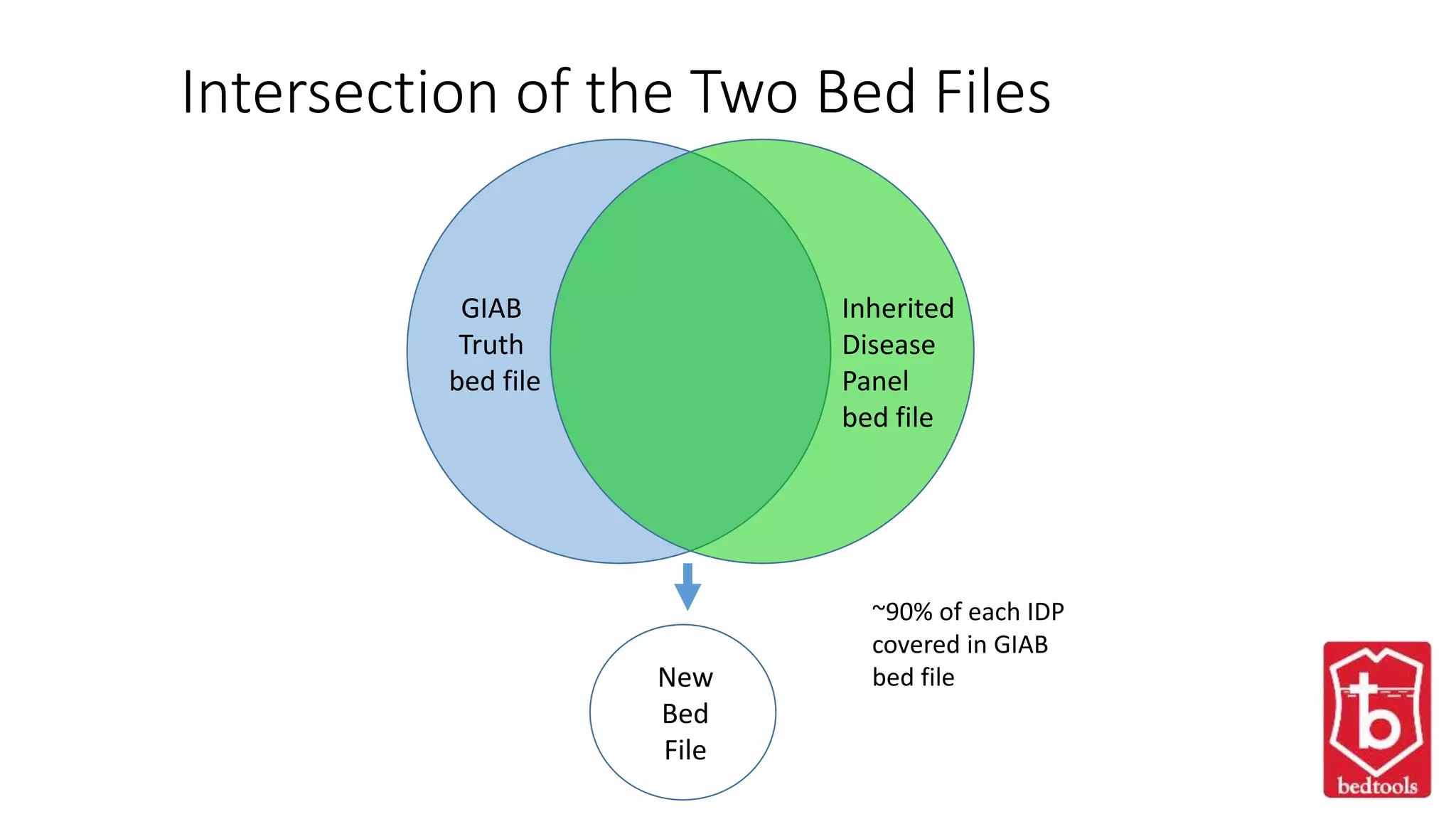
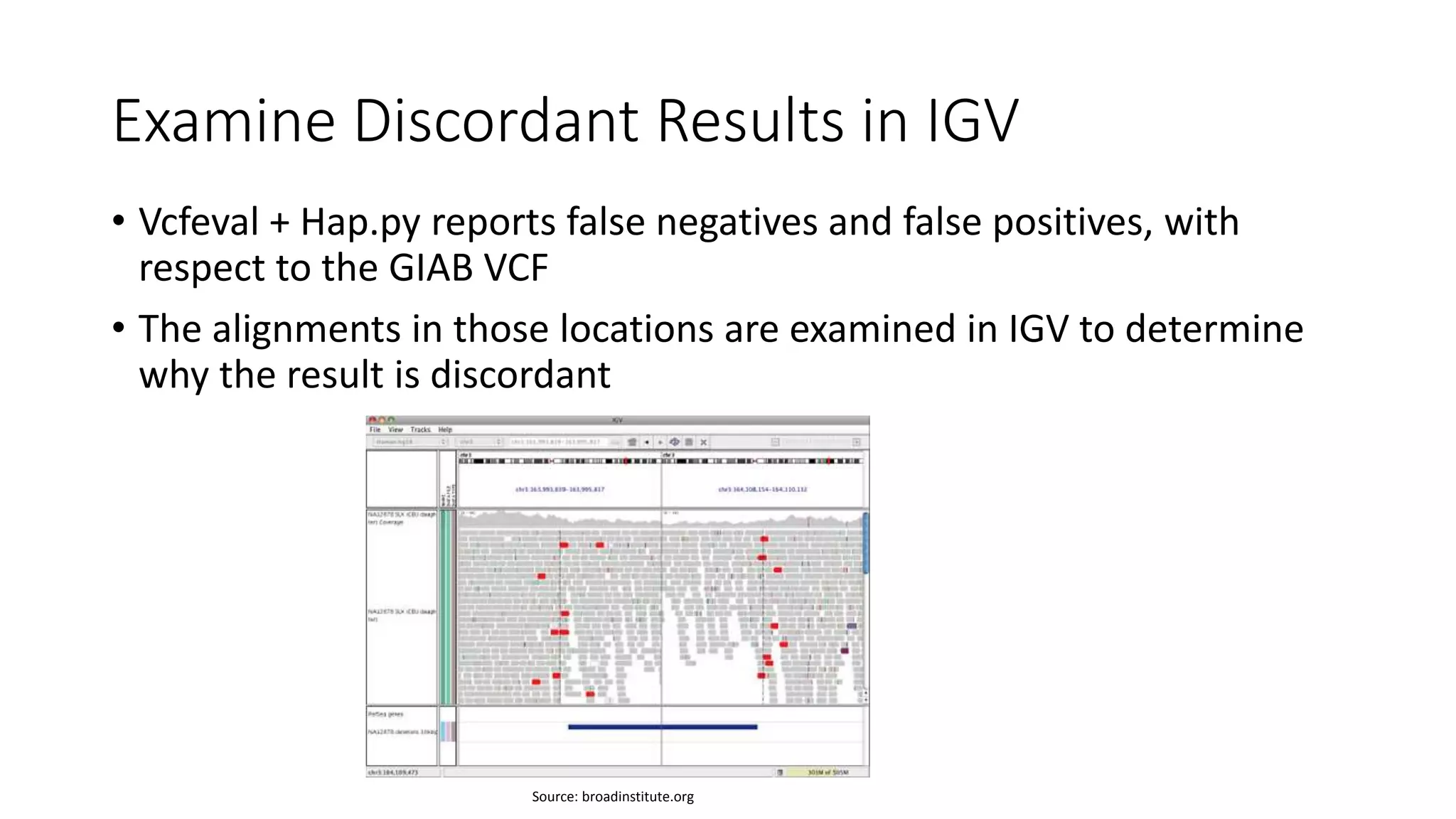
This document discusses targeted sequencing of Genome in a Bottle (GIAB) reference materials using commercial panels to further characterize the materials. Seven individuals from various GIAB materials will be sequenced in triplicate using various targeted panels, including cancer and inherited disease panels. The results will be compared to the high confidence GIAB regions to identify any discordant variants and potentially add new information. Overlap between different panels and to the GIAB regions will also be examined.