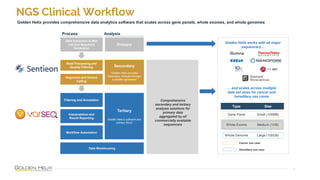
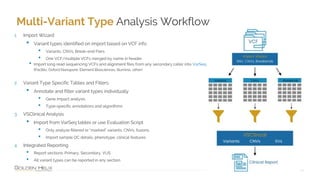
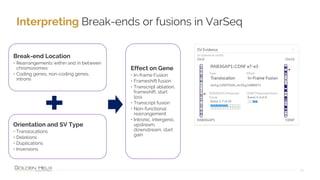
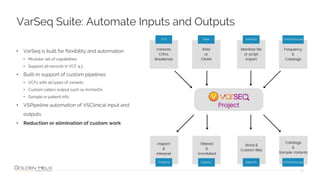
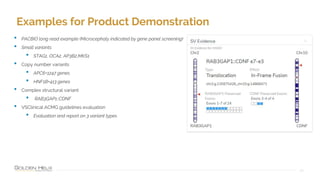
The document provides information about Golden Helix's VarSeq 2.4.0 software and its VSClinical ACMG workflow module. It summarizes a presentation given on the topic, including details about Golden Helix as a company, the content covered in the presentation, and examples that will be demonstrated including analysis of variants, copy number variants, and structural variants from PacBio long read sequencing data using VSClinical.