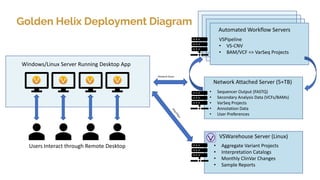
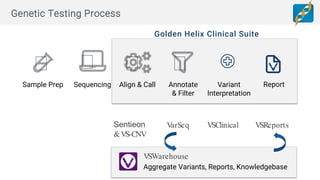
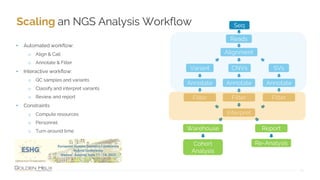
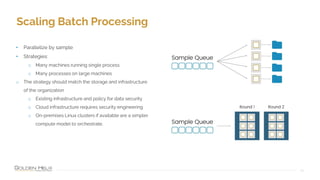
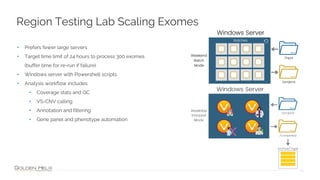
Golden Helix provides software solutions for clinical NGS workflows to meet scalability and data privacy requirements. Their software suite includes tools for variant calling, annotation, filtering, interpretation and reporting. They aim to help labs customize validated workflows to scale sample processing volume through batch processing strategies like parallelization across multiple machines. Golden Helix works with various customers globally, from regional testing labs to pharmaceutical companies, to develop scalable automated pipelines from panels to whole genomes.









![Pharma Services Scaling Genomes
16
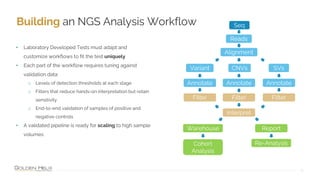
• Deliverables are locked down through versioned
pipelines
• All steps inside read-only Docker containers
• VSPipeline + Annotations + Project Templates
• Local Linux cluster and cloud deployment scenarios
• Secure Cloud deployments in China with no
outbound internet access
• Batch node job:
• Download docker containers
• [Cloud only] Download BAM, VCF from storage
• Docker run pipeline
• [Cloud only] Upload results to storage](https://image.slidesharecdn.com/2022-05-16evaluatingcloudvson-premisesforngsclinicalworkflowsv2-220518213451-c8b1beb5/85/Evaluating-Cloud-vs-On-Premises-for-NGS-Clinical-Workflows-16-320.jpg)