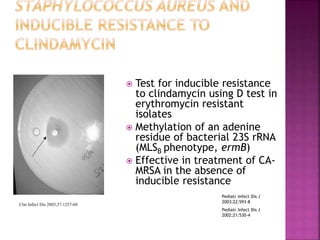
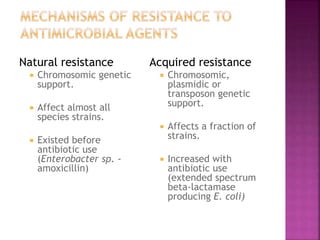
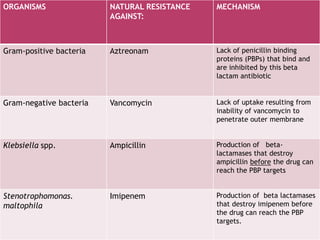
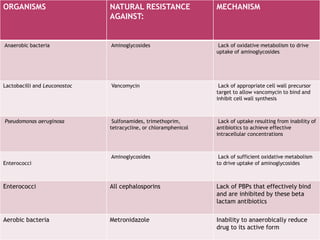
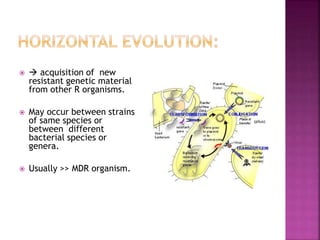
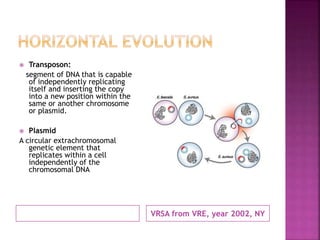
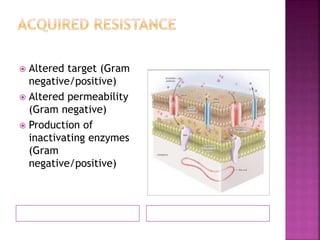
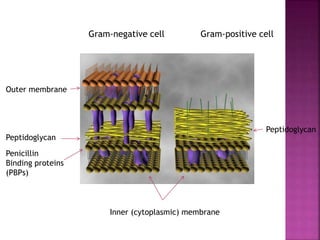
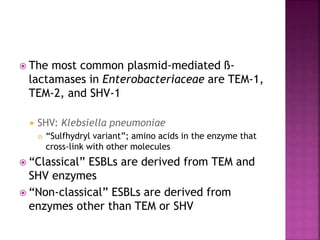
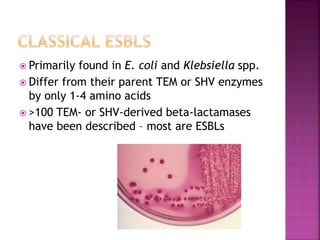
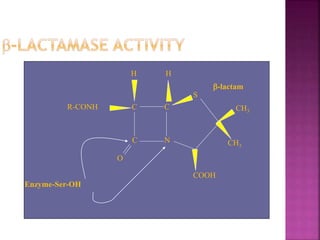
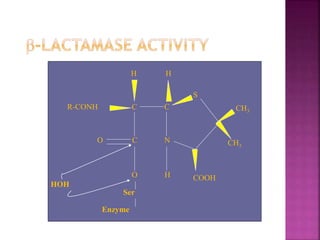
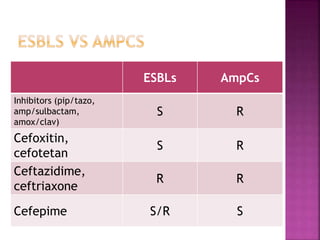
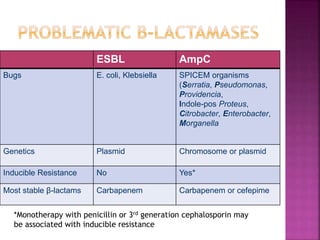
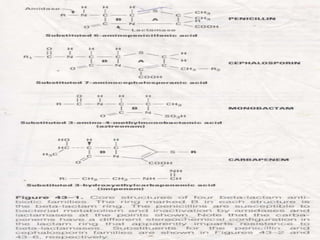
A 41-year-old woman with aplastic anemia was admitted with fever. Blood cultures grew E. coli resistant to ampicillin and narrow-spectrum cephalosporins. Despite treatment with multiple antimicrobials over 4 weeks, the patient's fever and bacteremia persisted. The microbiology lab was contacted to help determine why standard therapies were failing to clear the infection.












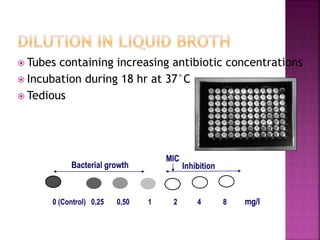
![ Minimum inhibitory concentration [MIC]
The smallest concentration of antibiotic that inhibits the
growth of organism.
Liquid media (dilution) allows MIC estimation
Solid media (diffusion)
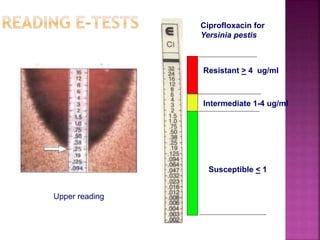
Disk diffusion (Kirby-Bauer)
E-tests
Allows MIC estimation
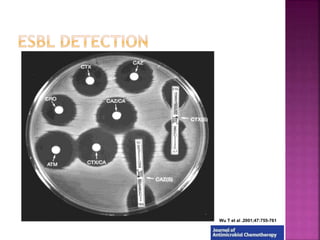
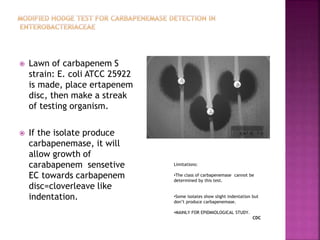
Beta lactamase production: quick screening method](https://image.slidesharecdn.com/antimicrobialresistance-151221083529/85/Antimicrobial-resistance-15-320.jpg)