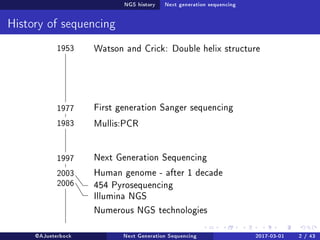
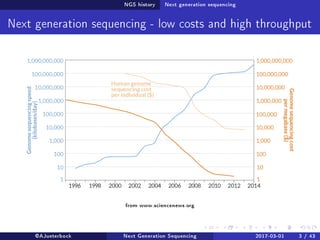
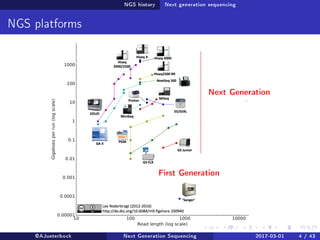
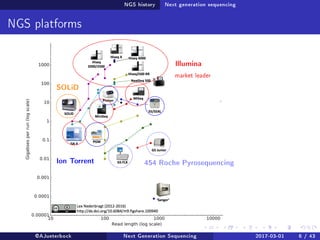
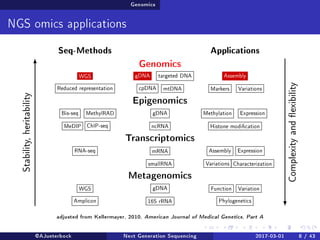
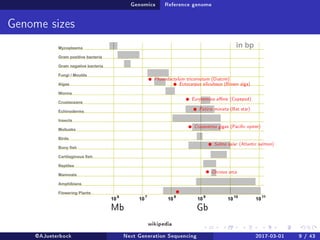
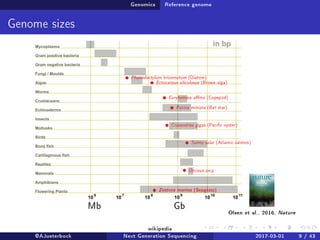
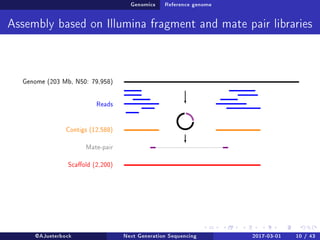
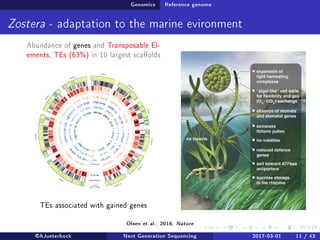
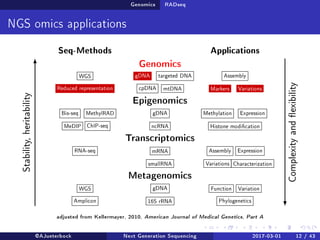
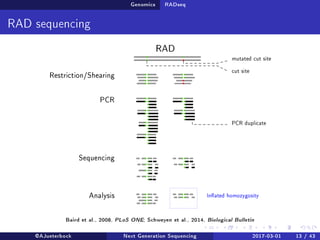
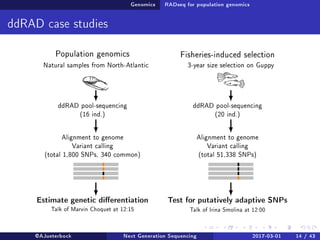
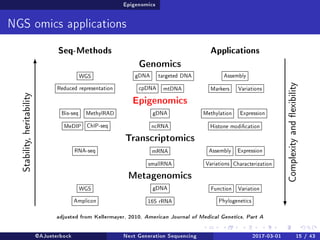
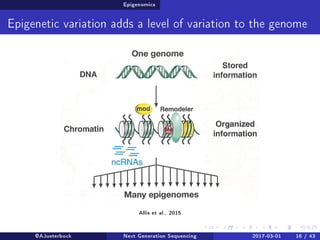
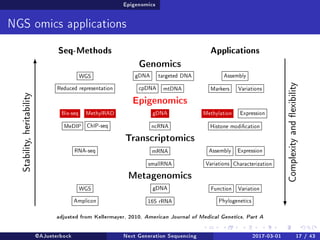
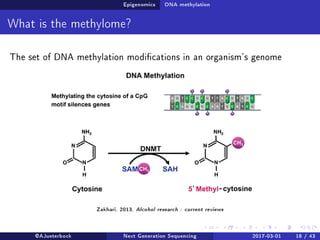
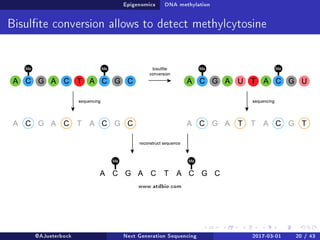
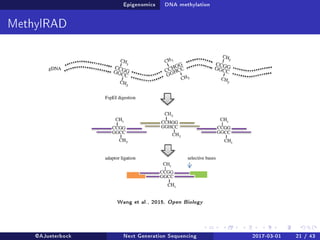
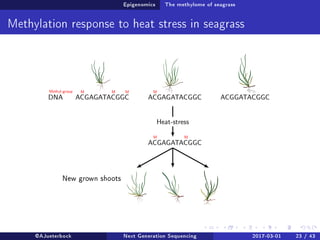
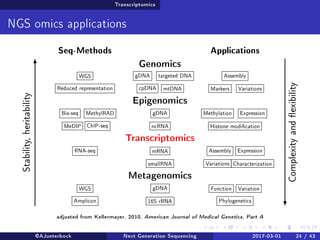
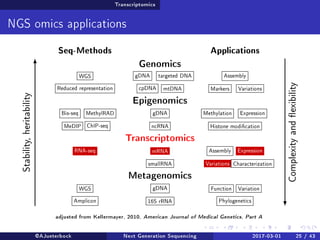
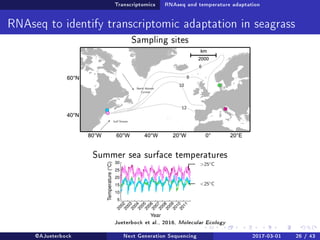
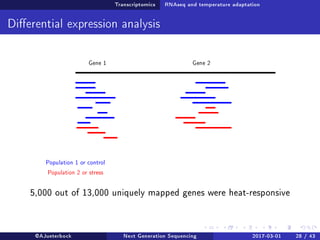
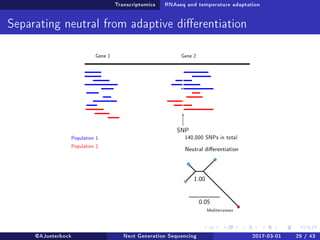
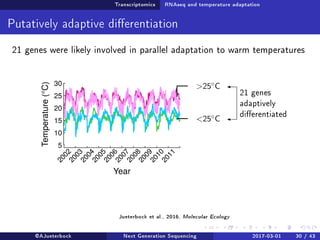
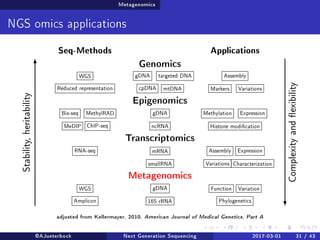
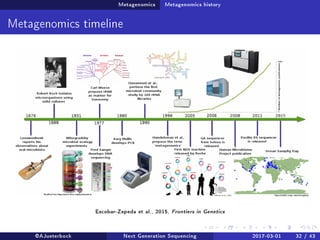
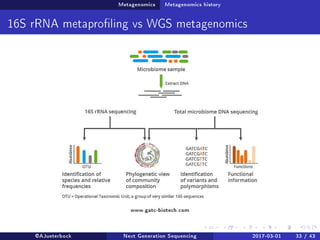
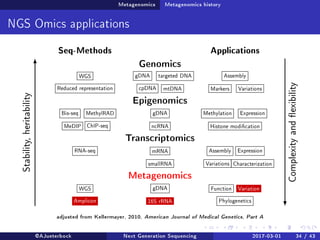
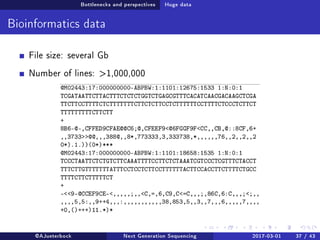
The document reviews the advancements and applications of next-generation sequencing (NGS) techniques in marine non-model organisms over the past decade. It highlights various sequencing technologies, their applications in genomics, transcriptomics, epigenomics, and metagenomics, as well as the challenges faced in data analysis and standardization. Additionally, it discusses the impact of third-generation sequencing technologies and the potential for innovations such as CRISPR genome editing.