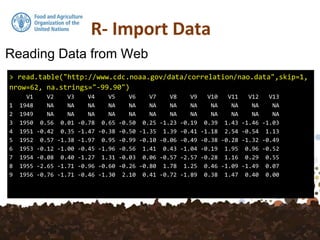
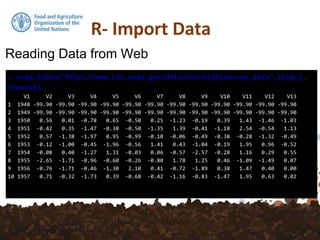
The document discusses various functions for reading data into R from files and databases, as well as writing data out of R. It describes read.csv and read.csv2 for reading comma separated value (CSV) files, with read.csv2 handling CSV formats with commas as decimal separators. It also covers reading from databases using functions like get_postgis_query and reading/writing directly from/to R using dump and source. Writing data out can be done with write.csv or to a format preserved R data with dump.








![> is.na(SOC)
Id UpperDepth LowerDepth SOC Lambda tsme Region
[1,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[2,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[3,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[4,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[5,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[6,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[7,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[8,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[9,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[10,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE
...](https://image.slidesharecdn.com/d3-1-r-data-import-data-export-170531132029/85/7-Data-Import-Data-Export-9-320.jpg)
![> anyNA(SOC)
[1] TRUE
> sum(is.na(SOC$SOC))
[1] 1](https://image.slidesharecdn.com/d3-1-r-data-import-data-export-170531132029/85/7-Data-Import-Data-Export-10-320.jpg)