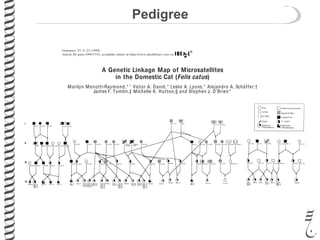
The document discusses a research project led by Hasan Alhaddad focusing on the genetic assessment of the Bengal breed of cats, particularly evaluating the genomic contributions of Asian leopard cats. It details three main aims: creating a diagnostic panel for identifying Asian leopard cat alleles, estimating the degree of 'bengalness' in various cat breeds, and understanding the Bengal breed's lineage. The findings highlight the genetic markers associated with Asian leopard cats and their implications for breeding and conservation efforts in feline genetics.