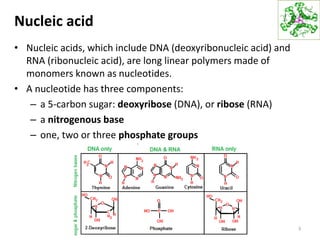
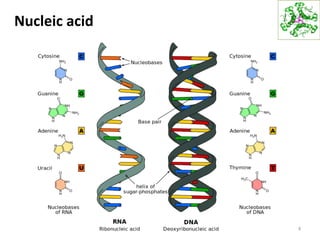
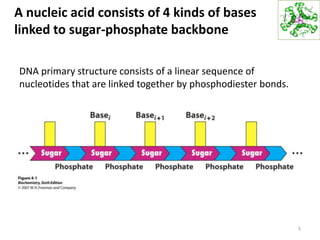
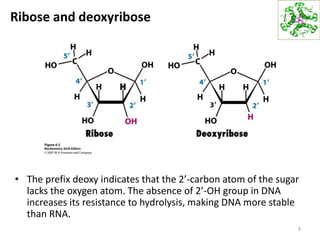
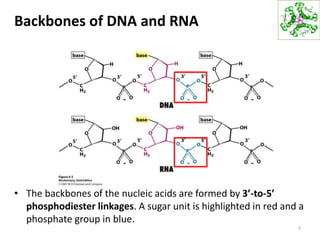
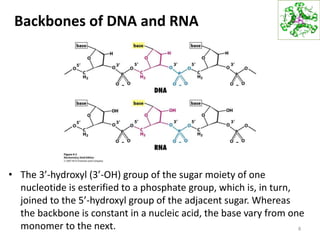
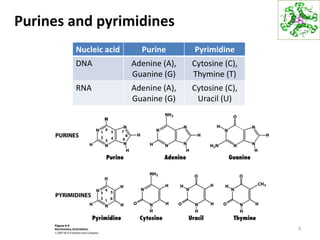
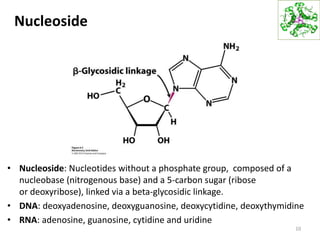
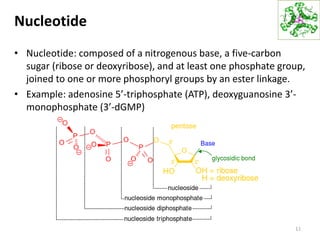
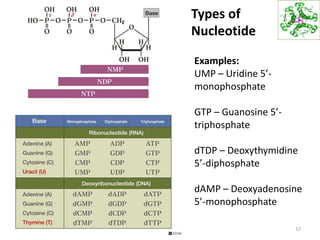
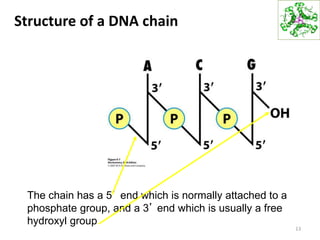
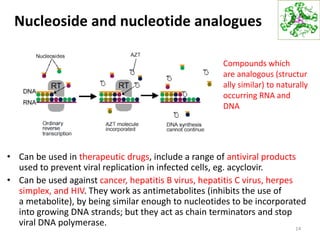
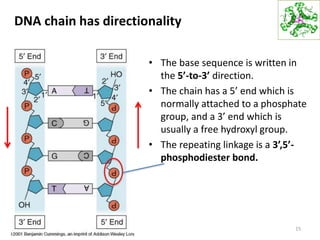
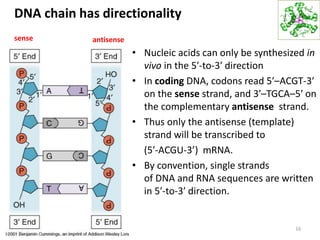
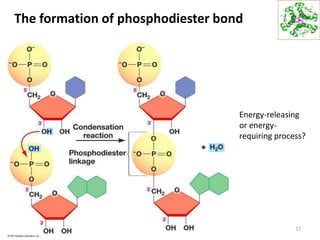
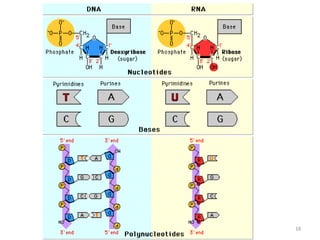
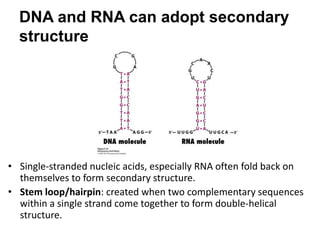
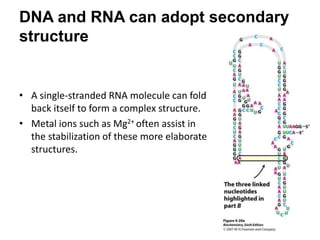
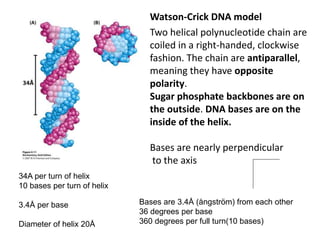
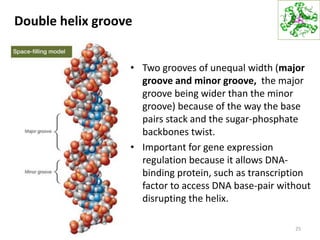
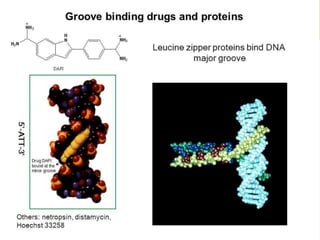
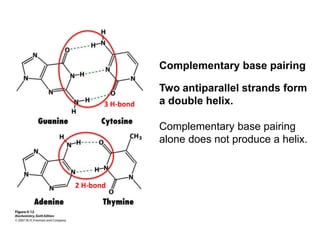
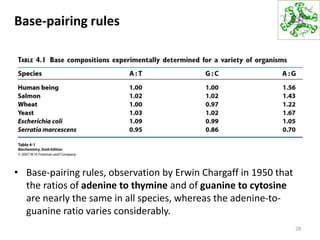
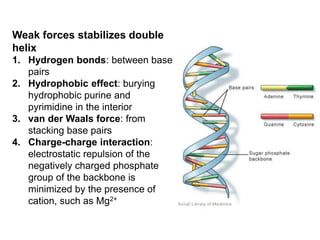
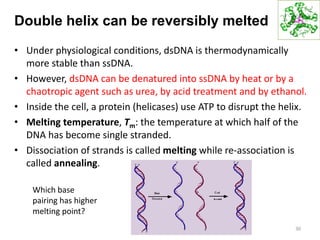
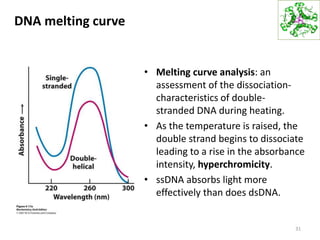
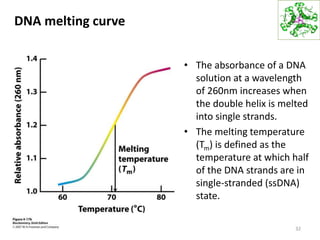
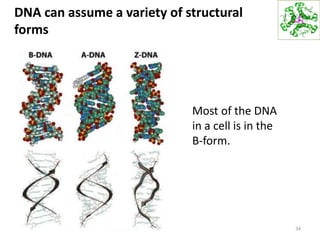
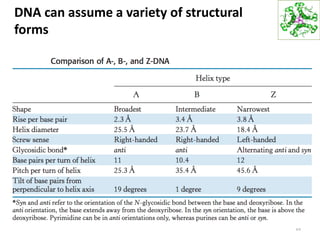
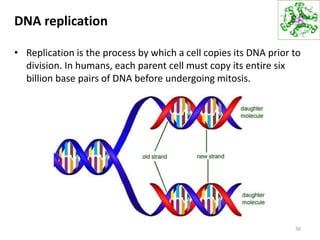
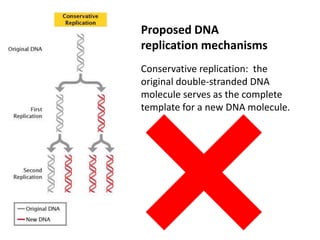
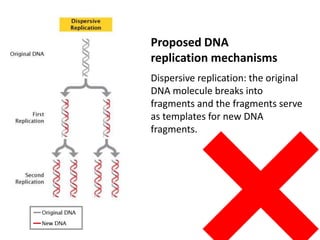
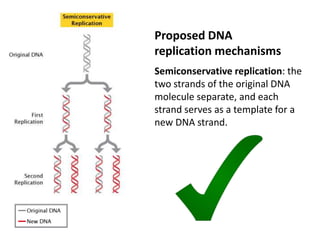
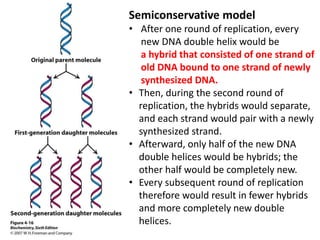
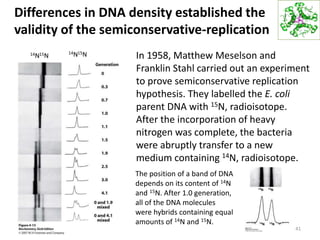
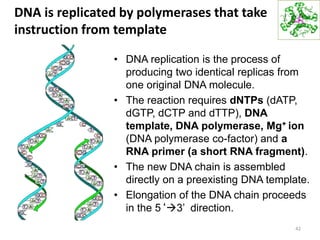
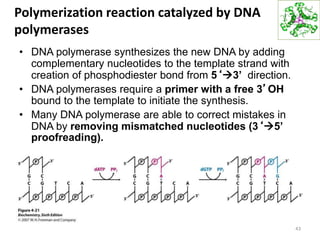
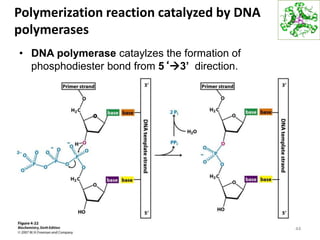
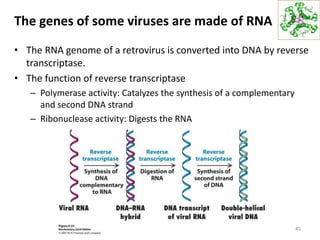
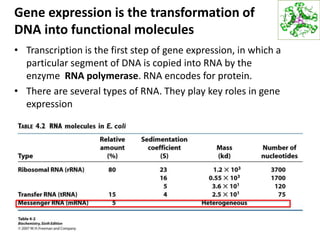
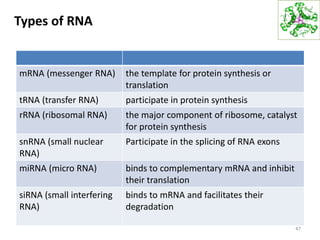
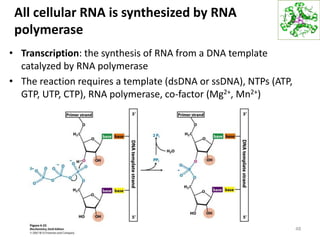
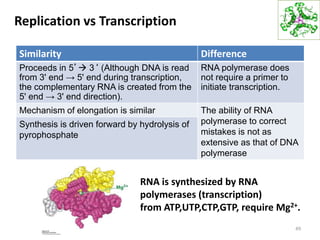
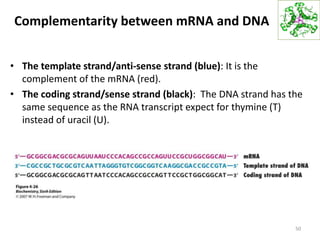
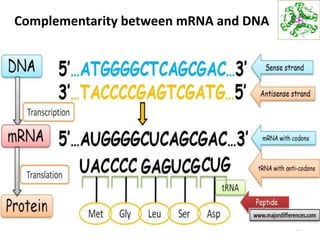
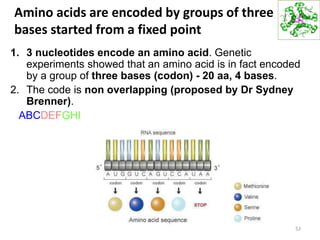
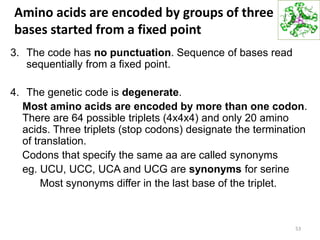
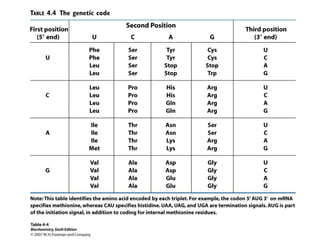
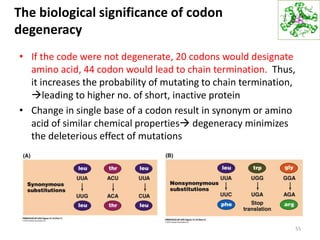
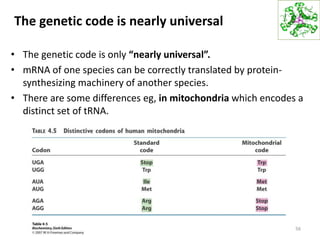
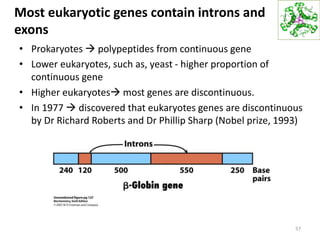
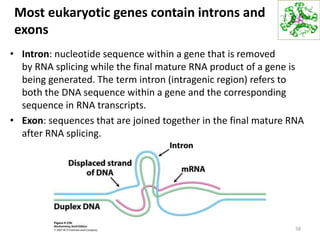
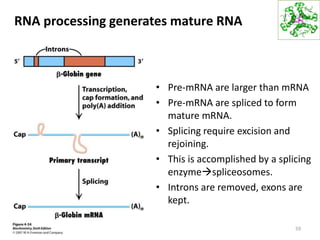
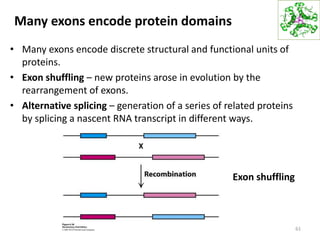
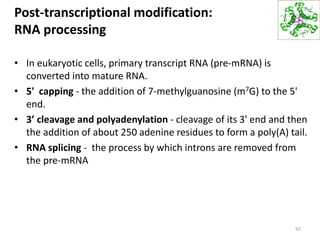
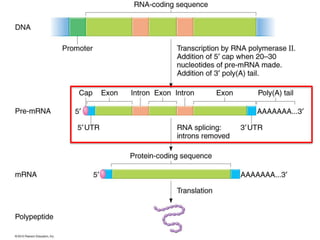
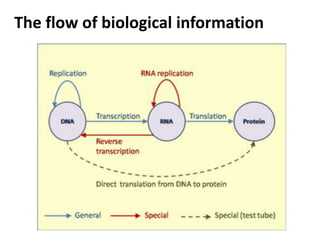
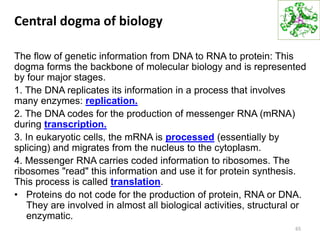
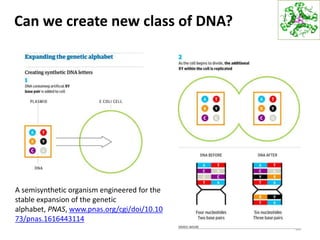
The document provides an overview of DNA, RNA, and the flow of genetic information. It describes the basic structures of nucleic acids including nucleotides, nucleosides, the sugar-phosphate backbone, and base pairing in DNA and RNA. It discusses the double helix structure of DNA proposed by Watson and Crick, including features such as directionality, grooves, and base pairing rules. It also covers DNA replication, noting that the semiconservative model in which each parental strand serves as a template for a new strand is accepted.