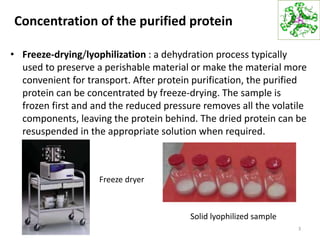
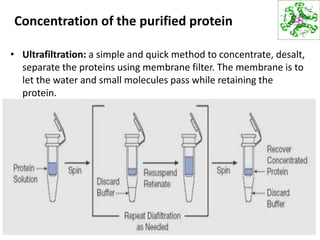
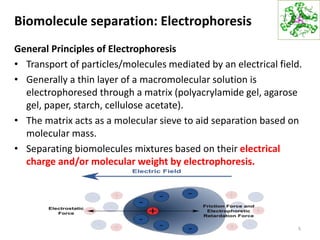
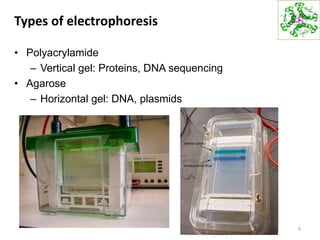
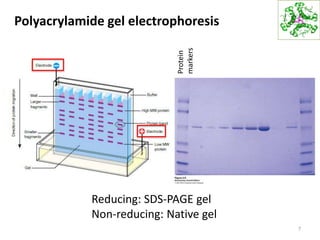
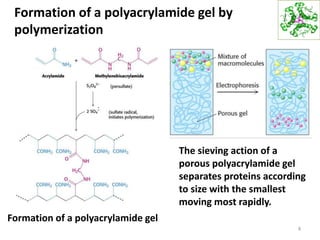
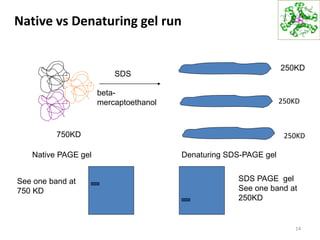
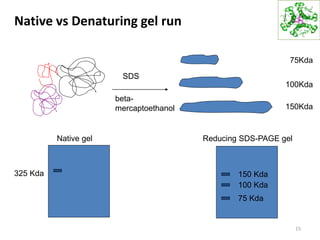
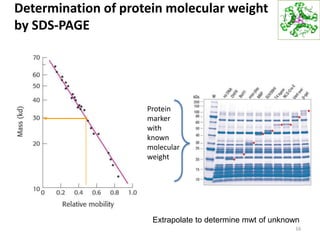
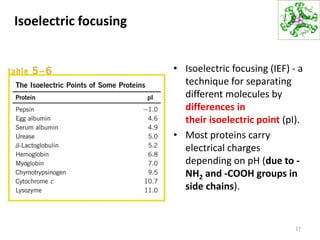
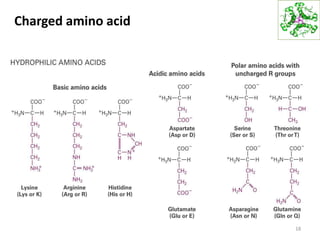
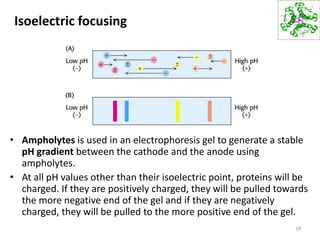
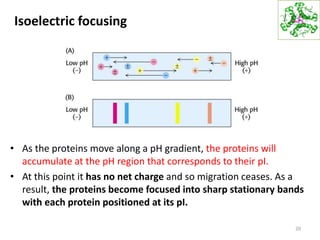
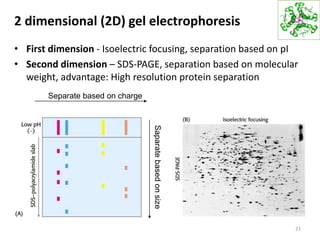
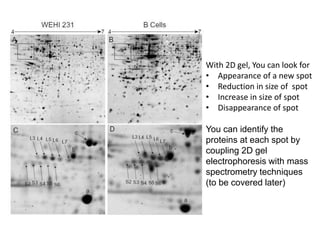
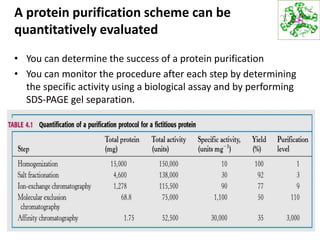
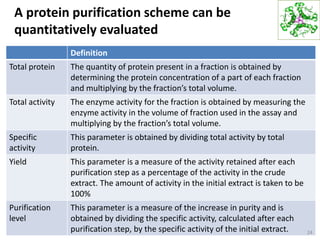
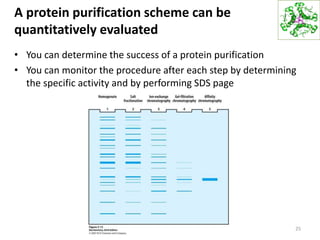
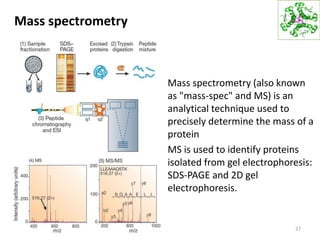
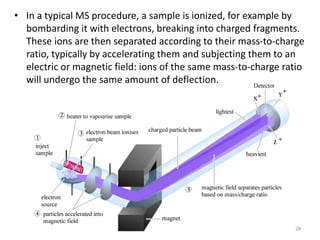
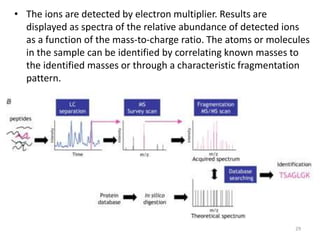
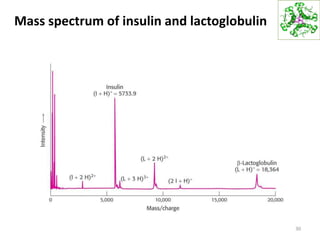
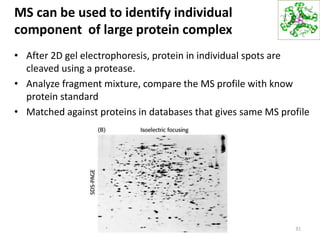
This document summarizes techniques for exploring and analyzing proteins, including concentrating purified proteins using lyophilization or ultrafiltration, separating proteins using electrophoresis or mass spectrometry, and identifying proteins using mass spectrometry. Electrophoresis techniques like SDS-PAGE and 2D gels separate proteins based on size and charge, allowing visualization and quantification of purified proteins. Mass spectrometry further identifies proteins by correlating detected ion masses with known protein standards. These techniques provide a quantitative evaluation of protein purification schemes.