The document discusses Bayesian neural networks and related topics. It covers Bayesian neural networks, stochastic neural networks, variational autoencoders, and modeling prediction uncertainty in neural networks. Key points include using Bayesian techniques like MCMC and variational inference to place distributions over the weights of neural networks, modeling both model parameters and predictions as distributions, and how this allows capturing uncertainty in the network's predictions.







![¤ +G
¤ #
¤
¤ +G
3G
$ +G ! = H $ +G # $ # ! 5# = IJ(A|K)[$ +G # ]
$ 3G +G, ! = H $ 3G +G, # $ # ! 5# = IJ(A|K)[$ 3G +G, # ]](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-8-320.jpg)
![¤
¤
¤
¤
¤
¤
$ +G ! = H $ +G # $ # ! 5# = IJ(A|K)[$ +G # ]
$ " ! =
$ ! " $(")
$(!)
$ # ! =
$ ! # $(#)
$(!)
=
$ ! # $(#)
∫ $ ! # $ # 5#](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-9-320.jpg)



![¤ # $(#)
¤ #
¤
Weight Uncertainty in Neural Networks
H1 H2 H3 1
X 1
Y
0.5 0.1 0.7 1.3
1.40.3
1.2
0.10.1 0.2
H1 H2 H3 1
X 1
Y
Figure 1. Left: each weight has a fixed value, as provided by clas-
sical backpropagation. Right: each weight is assigned a distribu-
tion, as provided by Bayes by Backprop.
is related to recent methods in deep, generative modelling
(Kingma and Welling, 2014; Rezende et al., 2014; Gregor
et al., 2014), where variational inference has been applied
to stochastic hidden units of an autoencoder. Whilst the
number of stochastic hidden units might be in the order of
the parameters of the categorical dis
through the exponential function then
regression Y is R and P(y|x, w) is a G
– this corresponds to a squared loss.
Inputs x are mapped onto the param
tion on Y by several successive layers
tion (given by w) interleaved with elem
transforms.
The weights can be learnt by maximum
tion (MLE): given a set of training exam
the MLE weights wMLE
are given by:
wMLE
= arg max
w
log P(D|w
= arg max
w
i
log P(
This is typically achieved by gradient
NN Bayesian NN
$(3G|!, +G, N) = H $ 3G +G, # $ # !, N 5#
[Blundell+ 2015]](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-13-320.jpg)





![¤ O(#|N)
¤ ST[O(#|N)||$ # ! ] N
¤ KL
¤ N ELBO N
→
ST[O(#|N)||$ (#|!)]
= −∫ O # N log
W K|A W A
J # N
5# + log $ !
= −ℒ !; N + log $ !
ELBO](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-23-320.jpg)
![ELBO
¤ ELBO ℒ !; N
1.
¤ $(#|!)
MC
¤
2. ELBO
¤ MC
¤ MC ∫ O # N log
W K|A W A
J # N
5# = IJ[log
W K|A W A
J # N
]
3.
¤ EM
¤ ELBO](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-24-320.jpg)

![¤ Practical Variational Inference for Neural Networks [Graves 2011]
¤
¤
¤ T9 O(#|N) N = {Z, [}
¤
ℒ !; N = ∫ O (#|N)log
$ !|# $(#)
O(#|N)
5#
= − EJ(A|])[log $(!|#)] + ST[O(#|N)||$(#)]
^T9
^Z
≈
1
a
B
^ log $(!|#)
^#
/
C
^T9
^[b
≈
1
2a
B
^ log $(!|#)
^#
b/
C](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-26-320.jpg)
![¤ Weight Uncertainty in Neural Networks [Blundell+ 2015]
¤
¤ O(#|N)
¤
¤ http://www.slideshare.net/masa_s/weight-uncertainty-in-neural-networks
Bayes by backprop
^
^N
ℒ !; N =
^
^N
IJ(A|]) d(#, N)
d #, N = log
$ !|# $(#)
O(#|N)
= IJ(e)
^d(#, N)
^N
^#
^N
+
^d(#, N)
^N
Reparameterization trick
# = Z + diag([) ⊙ i i~k(0, m)](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-27-320.jpg)
![Dropout
¤ Dropout as a Bayesian Approximation [Gal+ 2015]
¤ O # N = ∏ O(nC|oC)
¤ T9 = − EJ # N [log $(!|#)]
¤ oC 0
¤ 0
¤ dropout
¤ drop-connect multiplicative Gaussian noise
sults are summarised here
n uncertainty estimates for
el with L layers and a loss
max loss or the Euclidean
Wi the NN’s weight ma-
1, and by bi the bias vec-
ayer i = 1, ..., L. We de-
corresponding to input xi
the input and output sets
on a regularisation term is
egularisation weighted by
n a minimisation objective
λ
L
i=1
||Wi||2
2 + ||bi||2
2 .
(1)
variables for every input
in each layer (apart from
le takes value 1 with prob-
ropped (i.e. its value is set
orresponding binary vari-
me values in the backward
to the parameters.
p(y|x, ω) = N y; y(x, ω), τ ID
y x, ω = {W1, ...,WL}
=
1
KL
WLσ ...
1
K1
W2σ W1x + m1 ...
The posterior distribution p(ω|X, Y) in eq. (2) is in-
tractable. We use q(ω), a distribution over matrices whose
columns are randomly set to zero, to approximate the in-
tractable posterior. We define q(ω) as:
Wi = Mi · diag([zi,j]Ki
j=1)
zi,j ∼ Bernoulli(pi) for i = 1, ..., L, j = 1, ..., Ki−1
given some probabilities pi and matrices Mi as variational
parameters. The binary variable zi,j = 0 corresponds then
to unit j in layer i − 1 being dropped out as an input to
layer i. The variational distribution q(ω) is highly multi-
modal, inducing strong joint correlations over the rows of
the matrices Wi (which correspond to the frequencies in
the sparse spectrum GP approximation).
We minimise the KL divergence between the approximate
posterior q(ω) above and the posterior of the full deep GP,
p(ω|X, Y). This KL is our minimisation objective
− q(ω) log p(Y|X, ω)dω + KL(q(ω)||p(ω)). (3)](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-28-320.jpg)
![Dropout
¤ dropout
¤ dropout
¤
¤ MC dropout
¤ http://mlg.eng.cam.ac.uk/yarin/blog_2248.html
where ω = {Wi}L
i=1 is our set of random variables for a
model with L layers.
We will perform moment-matching and estimate the first
two moments of the predictive distribution empirically.
More specifically, we sample T sets of vectors of realisa-
tions from the Bernoulli distribution {zt
1, ..., zt
L}T
t=1 with
zt
i = [zt
i,j]Ki
j=1, giving {Wt
1, ..., Wt
L}T
t=1. We estimate
Eq(y∗|x∗)(y∗
) ≈
1
T
T
t=1
y∗
(x∗
, Wt
1, ..., Wt
L) (6)
following proposition C in the appendix. We refer to this
Monte Carlo estimate as MC dropout. In practice this
is equivalent to performing T stochastic forward passes
through the network and averaging the results.
This result has been presented in the literature before as
model averaging. We have given a new derivation for this
result which allows us to derive mathematically grounded
uncertainty estimates as well. Srivastava et al. (2014, sec-
tion 7.5) have reasoned empirically that MC dropout can
be approximated by averaging the weights of the network
(multiplying each Wi by pi at test time, referred to as stan-
dard dropout).
We estimate the second raw moment in the same way:
log p(y∗
|x∗
, X, Y) ≈
with a log-sum-exp o
passes through the ne
Our predictive distr
highly multi-modal,
give a glimpse into i
proximating variation
matrix column is bi-
tribution over each la
3.2 in the appendix).
Note that the dropo
To estimate the predi
we simply collect the
through the model.
used with existing N
thermore, the forward
sulting in constant run
dropout.
5. Experiments
T t=1
following proposition C in the appendix. We refer to this
Monte Carlo estimate as MC dropout. In practice this
is equivalent to performing T stochastic forward passes
through the network and averaging the results.
This result has been presented in the literature before as
model averaging. We have given a new derivation for this
result which allows us to derive mathematically grounded
uncertainty estimates as well. Srivastava et al. (2014, sec-
tion 7.5) have reasoned empirically that MC dropout can
be approximated by averaging the weights of the network
(multiplying each Wi by pi at test time, referred to as stan-
dard dropout).
We estimate the second raw moment in the same way:
Eq(y∗|x∗) (y∗
)T
(y∗
) ≈ τ−1
ID
+
1
T
T
t=1
y∗
(x∗
, Wt
1, ..., Wt
L)T
y∗
(x∗
, Wt
1, ..., Wt
L)
following proposition D in the appendix. To obtain the
model’s predictive variance we have:
Varq(y∗|x∗) y∗
≈ τ−1
ID
2
In the appendix (section 4.1) we extend this derivation to
classification. E(·) is defined as softmax loss and τ is set to 1.
proximating variational distributio
matrix column is bi-modal, and
tribution over each layer’s weight
3.2 in the appendix).
Note that the dropout NN mod
To estimate the predictive mean a
we simply collect the results of s
through the model. As a result,
used with existing NN models tra
thermore, the forward passes can
sulting in constant running time id
dropout.
5. Experiments
We next perform an extensive ass
of the uncertainty estimates obta
and convnets on the tasks of regr
We compare the uncertainty obtai
architectures and non-linearities,
olation, and show that model unc
classification tasks using MNIST
as an example. We then show th
tainty we can obtain a considerabl
tive log-likelihood and RMSE co
of-the-art methods. We finish wi](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-29-320.jpg)
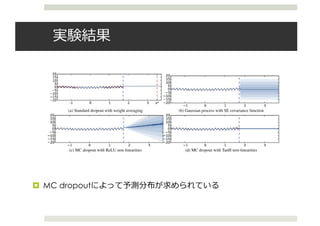
![Dropout
¤ Variational dropout and the local reparameterization trick
[Kingma+ 2015]
¤
¤ 0
¤ 0 local reparameterization
trick
¤ Dropout
¤
¤ http://www.slideshare.net/masa_s/dl-hacks-variational-dropout-and-the-local-
reparameterization-trick
2.2 Variance of the SGVB estimator
The theory of stochastic approximation tells us that stochastic gradient ascent using (3) will asymp-
totically converge to a local optimum for an appropriately declining step size and sufficient weight
updates [18], but in practice the performance of stochastic gradient ascent crucially depends on
the variance of the gradients. If this variance is too large, stochastic gradient descent will fail
to make much progress in any reasonable amount of time. Our objective function consists of an
expected log likelihood term that we approximate using Monte Carlo, and a KL divergence term
DKL(qφ(w)||p(w)) that we assume can be calculated analytically and otherwise be approximated
with Monte Carlo with similar reparameterization.
Assume that we draw minibatches of datapoints with replacement; see appendix F for a similar
analysis for minibatches without replacement. Using Li as shorthand for log p(yi
|xi
, w = f(ϵi
, φ)),
the contribution to the likelihood for the i-th datapoint in the minibatch, the Monte Carlo estimator
(3) may be rewritten as LSGVB
D (φ) = N
M
M
i=1 Li, whose variance is given by
Var LSGVB
D (φ) =
N2
M2
M
i=1
Var [Li] + 2
M
i=1
M
j=i+1
Cov [Li, Lj] (4)
=N2 1
M
Var [Li] +
M − 1
M
Cov [Li, Lj] , (5)
where the variances and covariances are w.r.t. both the data distribution and ϵ distribution, i.e.
Var [Li] = Varϵ,xi,yi log p(yi
|xi
, w = f(ϵ, φ)) , with xi
, yi
drawn from the empirical distribu-
tion defined by the training set. As can be seen from (5), the total contribution to the variance by
Var [Li] is inversely proportional to the minibatch size M. However, the total contribution by the
covariances does not decrease with M. In practice, this means that the variance of LSGVB
D (φ) can be
dominated by the covariances for even moderately large M.
2.3 Local Reparameterization Trick
We therefore propose an alternative estimator for which we have Cov [Li, Lj] = 0, so that the vari-
ance of our stochastic gradients scales as 1/M. We then make this new estimator computationally
efficient by not sampling ϵ directly, but only sampling the intermediate variables f(ϵ) through which
SGVB](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-31-320.jpg)


![Automatic Differentiation
Variational Inference ADVI
¤ Automatic Differentiation Variational Inference [Kucukelbir+ 2016]
¤ Stan PyMC3 Edward
¤ ADVI
1. N $(+, N) $(+, p)
ST[O(p)||$(p, +))]
2. O MC
3. O](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-34-320.jpg)


![¤
¤
¤
¤ L
¤
¤
.4 Variational Approximations in Real Coordinate Space
fter the transformation, the latent variables ζ have support in the real coordinate space K
. We hav
choice of variational approximations in this space. Here, we consider Gaussian distributions; thes
mplicitly induce non-Gaussian variational distributions in the original latent variable space.
Mean-field Gaussian. One option is to posit a factorized (mean-field) Gaussian variational approxima
on
q(ζ; φ) = ζ; µ,diag(σ2
) =
K
k=1
ζk ; µk,σ2
k ,
where the vector φ = (µ1,··· ,µK ,σ2
1,··· ,σ2
K ) concatenates the mean and variance of each Gaussia
actor. Since the variance parameters must always be positive, the variational parameters live in th
et Φ = { K
, K
>0}. Re-parameterizing the mean-field Gaussian removes this constraint. Consider th
4
Stan provides various transformations for upper and lower bounds, simplex and ordered vectors, and structured matrices suc
covariance matrices and Cholesky factors.
6
N
x [ n ] ~ poisson ( theta ) ;
}
Figure 2: Specifying a simple nonconjugate probability model in Stan.
Figure 2: Specifying a simple nonconjugate probability model in Stan.
arithm of the standard deviations, ω = log(σ), applied element-wise. The support of ω is n
real coordinate space and σ is always positive. The mean-field Gaussian becomes q(ζ; φ)
ζ; µ,diag(exp(ω)2
) , where the vector φ = (µ1,··· ,µK ,ω1,··· ,ωK ) concatenates the mean a
arithm of the standard deviation of each factor. Now, the variational parameters are unconstrain
2K
.
l-rank Gaussian. Another option is to posit a full-rank Gaussian variational approximation
q(ζ; φ) = ζ; µ,Σ ,
ere the vector φ = (µ,Σ) concatenates the mean vector µ and covariance matrix Σ. To ensure t
always remains positive semidefinite, we re-parameterize the covariance matrix using a Chole
torization, Σ = LL⊤
. We use the non-unique definition of the Cholesky factorization where
gonal elements of L need not be positively constrained (Pinheiro and Bates, 1996). Therefor
s in the unconstrained space of lower-triangular matrices with K(K + 1)/2 real-valued entries. T
-rank Gaussian becomes q(ζ; φ) = ζ; µ, LL⊤
, where the variational parameters φ = (µ, L)
constrained in K+K(K+1)/2
.
Figure 2: Specifying a simple nonconjugate probability model in Stan.
Figure 2: Specifying a simple nonconjugate probability model in Stan.
logarithm of the standard deviations, ω = log(σ), applied element-wise. The support of ω
the real coordinate space and σ is always positive. The mean-field Gaussian becomes q(ζ
ζ; µ,diag(exp(ω)2
) , where the vector φ = (µ1,··· ,µK ,ω1,··· ,ωK ) concatenates the m
logarithm of the standard deviation of each factor. Now, the variational parameters are uncon
in 2K
.
Full-rank Gaussian. Another option is to posit a full-rank Gaussian variational approximation
q(ζ; φ) = ζ; µ,Σ ,
where the vector φ = (µ,Σ) concatenates the mean vector µ and covariance matrix Σ. To ens
Σ always remains positive semidefinite, we re-parameterize the covariance matrix using a C
factorization, Σ = LL⊤
. We use the non-unique definition of the Cholesky factorization wh
diagonal elements of L need not be positively constrained (Pinheiro and Bates, 1996). The
lives in the unconstrained space of lower-triangular matrices with K(K + 1)/2 real-valued entr
full-rank Gaussian becomes q(ζ; φ) = ζ; µ, LL⊤
, where the variational parameters φ = (µ
unconstrained in K+K(K+1)/2
.
The full-rank Gaussian generalizes the mean-field Gaussian approximation. The off-diagonal term
covariance matrix Σ capture posterior correlations across latent random variables.5
This leads to
accurate posterior approximation than the mean-field Gaussian; however, it comes at a compu
cost. Various low-rank approximations to the covariance matrix reduce this cost, yet limit its a](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-37-320.jpg)

![¤ Black-box variational inference [Ranganath+ 2014]
¤
¤ ADVI likelihood ratio trick
¤ reparameterization trick
3.2 Variance of the Stochastic Gradients
ADVI uses Monte Carlo integration to approximate gradients of the ELBO, and then uses these gradients
in a stochastic optimization algorithm (Section 2). The speed of ADVI hinges on the variance of the
gradient estimates. When a stochastic optimization algorithm suffers from high-variance gradients, it
must repeatedly recover from poor parameter estimates.
ADVI is not the only way to compute Monte Carlo approximations of the gradient of the ELBO. Black
box variational inference (BBVI) takes a different approach (Ranganath et al., 2014). The BBVI gradient
estimator uses the gradient of the variational approximation and avoids using the gradient of the model.
For example, the following BBVI estimator
∇BBVI
µ = q(ζ;φ) ∇µ logq(ζ; φ) log p x, T−1
(ζ) + log det JT−1 (ζ) − logq(ζ; φ)
and the ADVI gradient estimator in Equation (7) both lead to unbiased estimates of the exact gradient.
While BBVI is more general—it does not require the gradient of the model and thus applies to more
settings—its gradients can suffer from high variance.
Figure 8 empirically compares the variance of both estimators for two models. Figure 8a shows the vari-
ance of both gradient estimators for a simple univariate model, where the posterior is a Gamma(10,10).
We estimate the variance using ten thousand re-calculations of the gradient ∇φ , across an increasing
number of MC samples M. The ADVI gradient has lower variance; in practice, a single sample suffices.
(See the experiments in Section 4.)
Figure 8b shows the same calculation for a 100-dimensional nonlinear regression model with likeli-
hood (y | tanh(x⊤
β), I) and a Gaussian prior on the regression coefficients β. Because this is a
multivariate example, we also show the BBVI gradient with a variance reduction scheme using control
variates described in Ranganath et al. (2014). In both cases, the ADVI gradients are statistically more
efficient.
100
101
102
103
100
101
102
103
Number of MC samples
Variance
(a) Univariate Model
100
101
102
103
10−3
10−1
101
103
Number of MC samples
ADVI
BBVI
BBVI with
control variate
(b) Multivariate Nonlinear Regression Model
Figure 8: Comparison of gradient estimator variances. The ADVI gradient estimator exhibits lower
variance than the BBVI estimator. Moreover, it does not require control variate variance reduction, which
is not available in univariate situations.
Figure 8: Comparison of gradient estimator variances. The ADVI gradient estimator exhibits lower
variance than the BBVI estimator. Moreover, it does not require control variate variance reduction, which
is not available in univariate situations.](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-39-320.jpg)


![¤ Stan
¤
¤ MCMC NUTS[Hoffman+ 2014] HMC
¤ Stan python R
¤ ADVI
¤ PyMC3
¤ Python MCMC
¤ Theano GPU
¤ ADVI
¤ Edward
¤
¤ criticism
¤ Python Tensorflow Keras
¤ Stan PyMC3 35x [Tran+ 2016]](https://image.slidesharecdn.com/20170127-170130120133/85/DL-hacks-Bayesian-Neural-Network-42-320.jpg)
