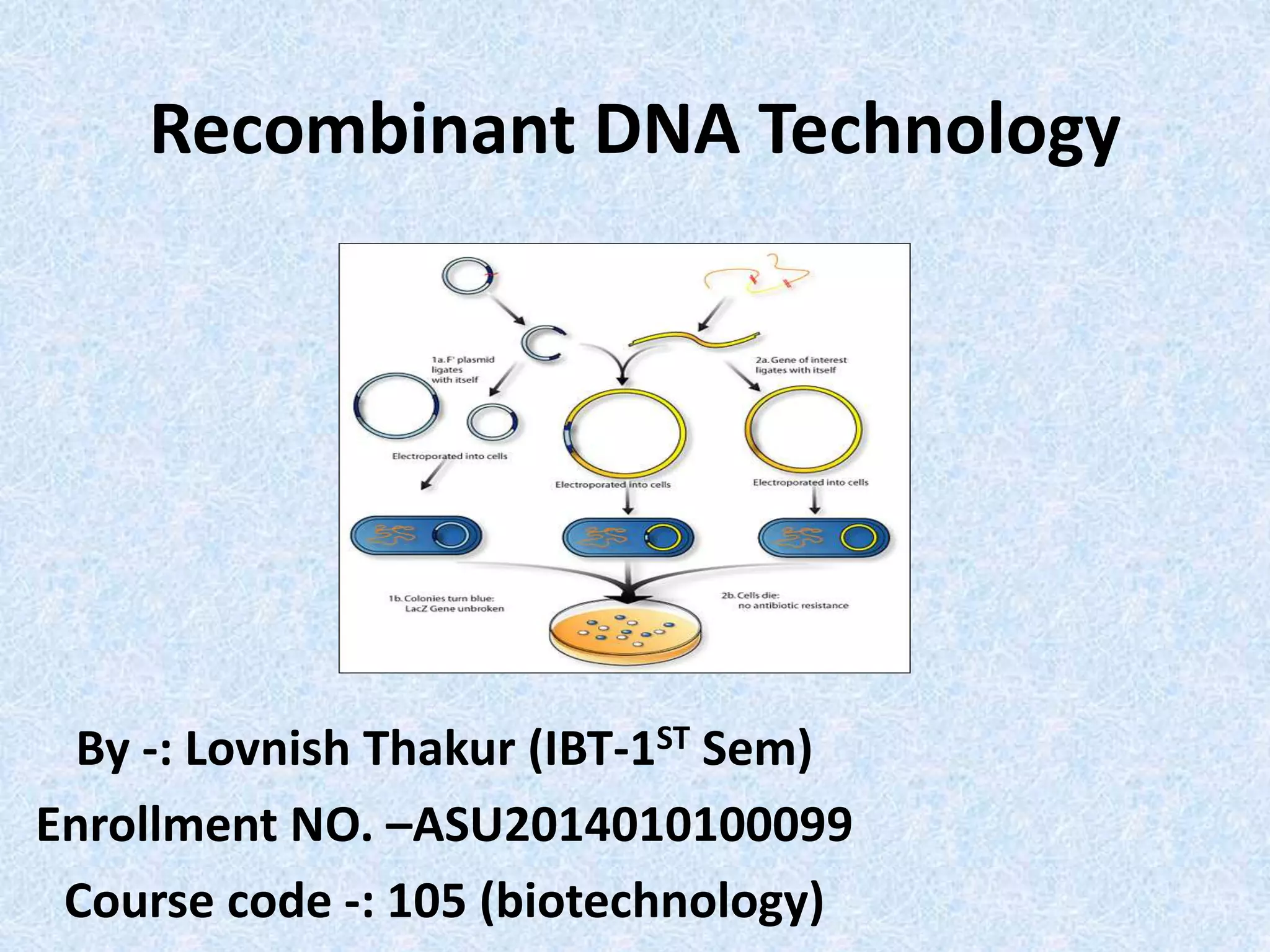
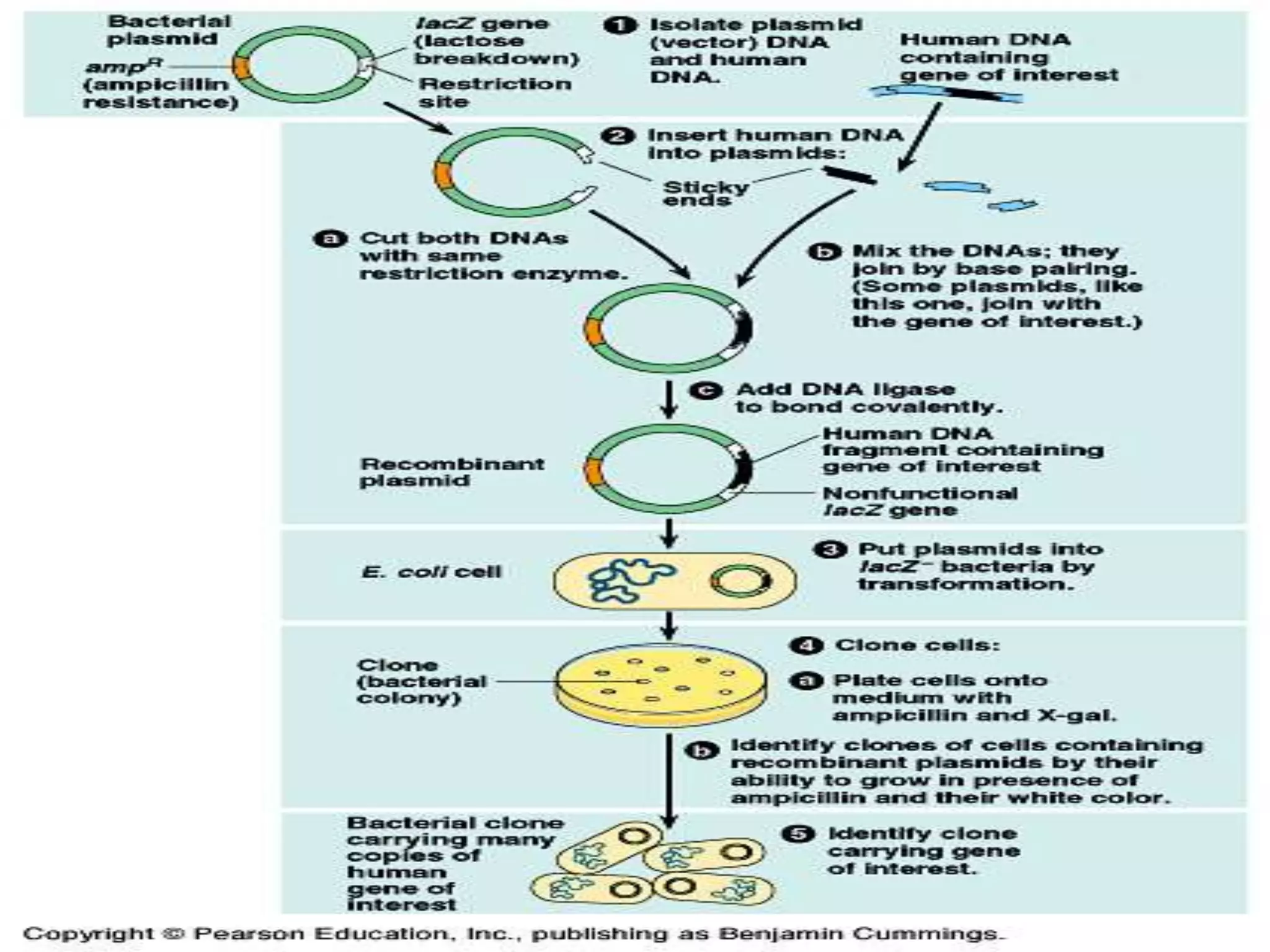
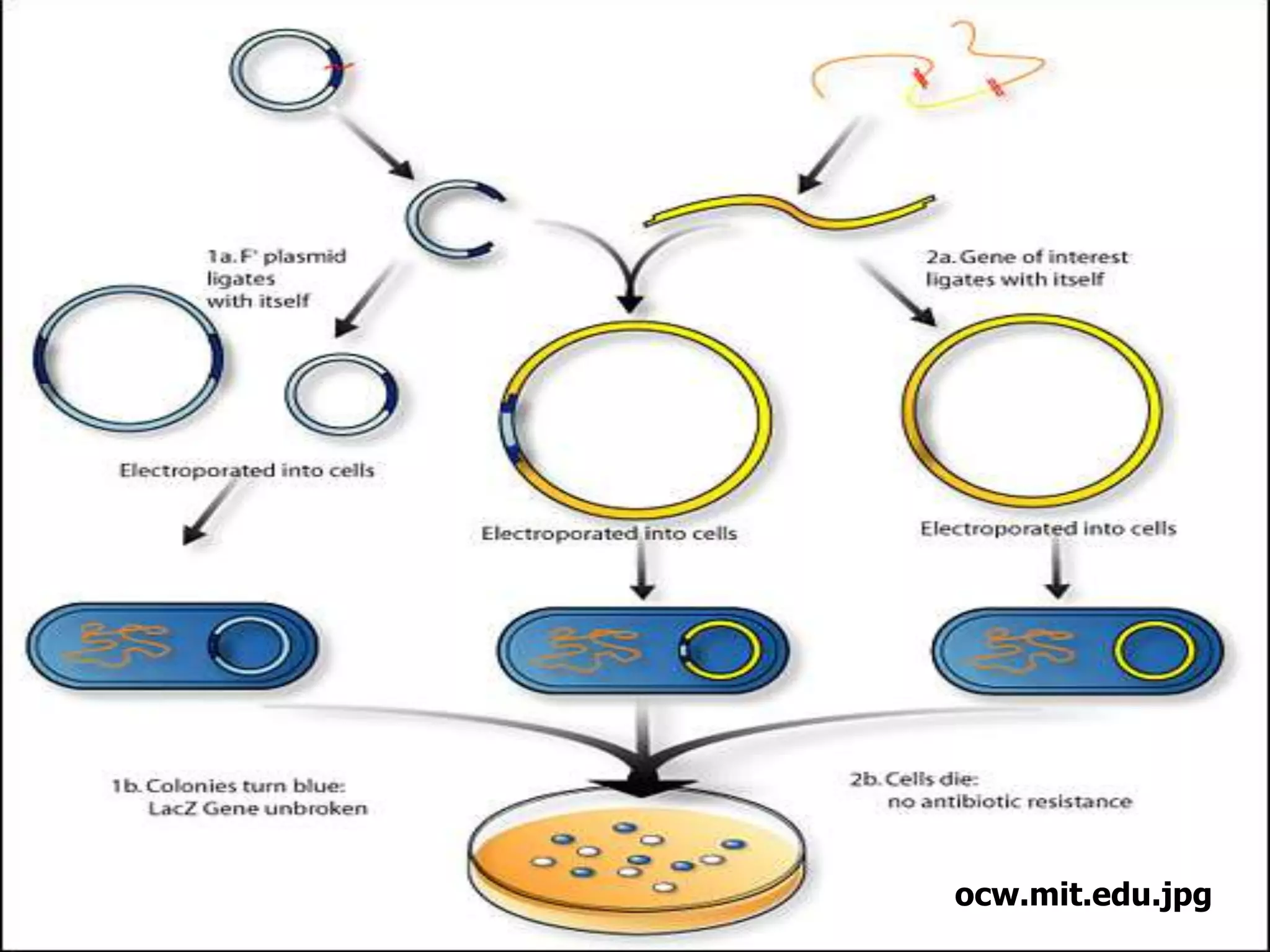
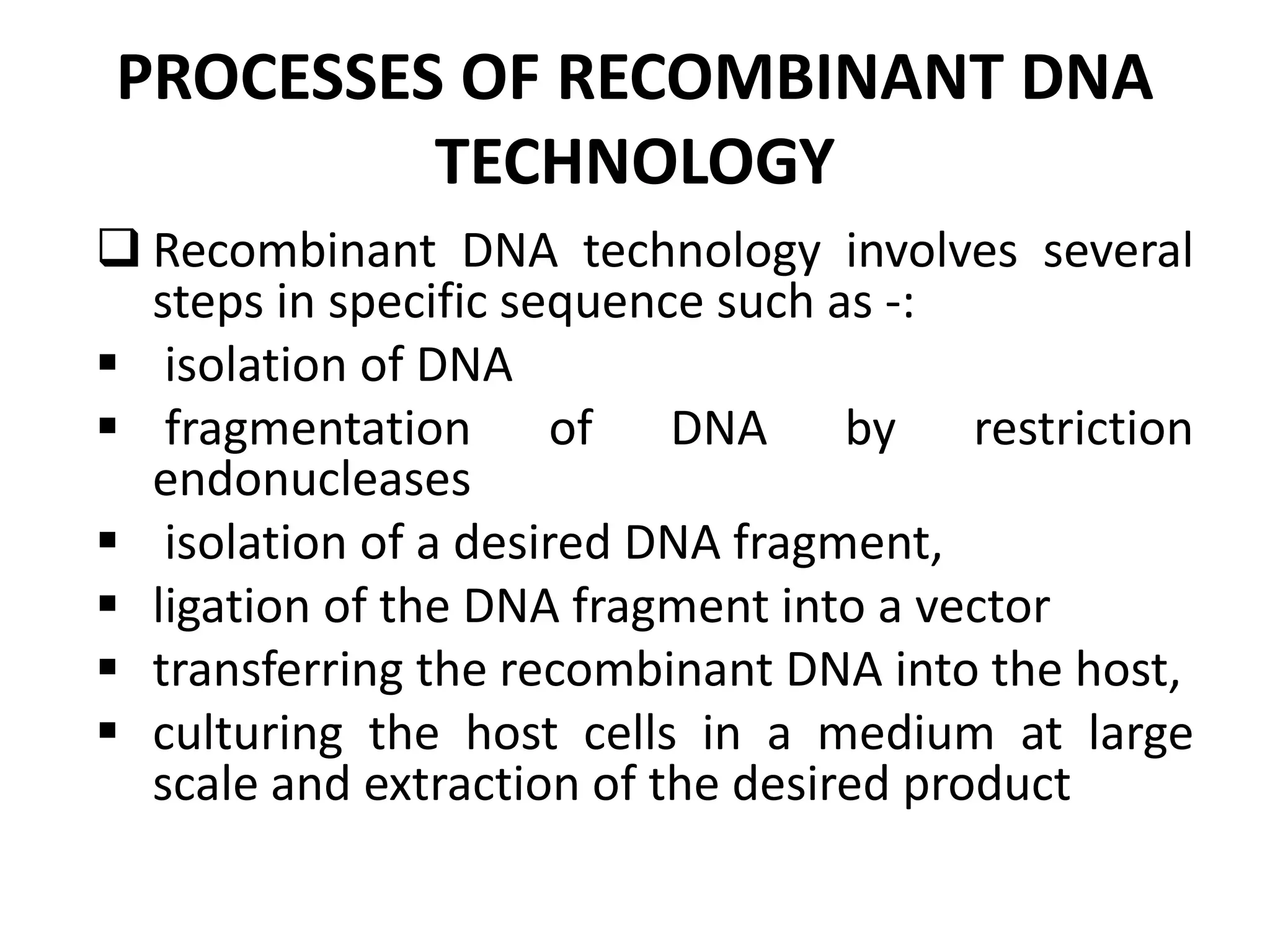
Recombinant DNA technology allows DNA from different species to be isolated, cut, and spliced together to form new recombinant molecules. Key tools for recombinant DNA technology include restriction enzymes, ligases, polymerases, vectors, and host cells. Recombinant DNA technology has many applications, including producing human insulin and other proteins for medical use, genetically engineering plants for crop improvement, and DNA fingerprinting for criminal investigations.