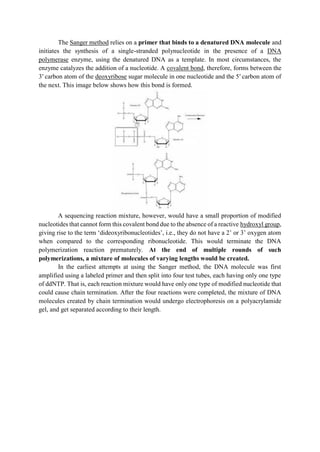
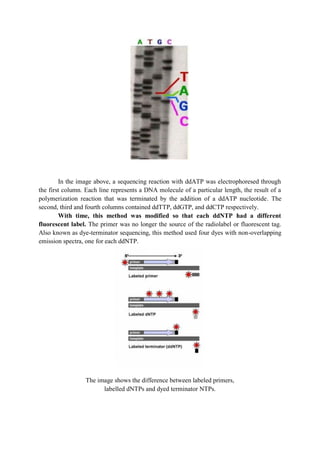
The document discusses DNA sequencing, a process for determining the nucleotide sequence of DNA, which aids in understanding genetic relationships among organisms. It outlines traditional and modern sequencing methods, including the Sanger method and high-throughput sequencing, highlighting their applications in genomics, diagnostics, molecular biology, and forensics. Advances in technology have reduced sequencing time and costs, making it integral for various scientific research areas and medical diagnoses.