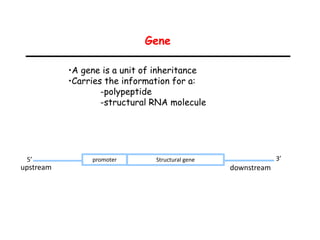
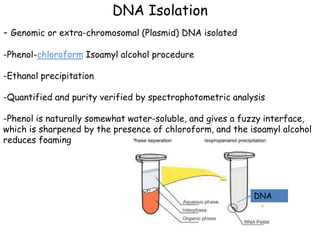
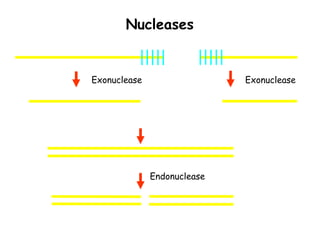
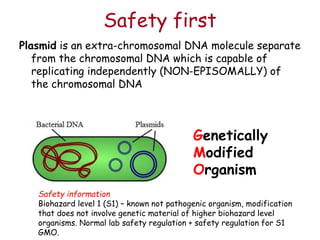
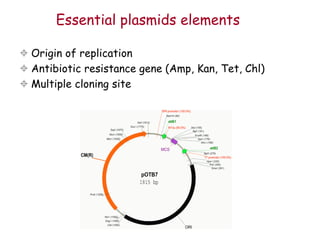
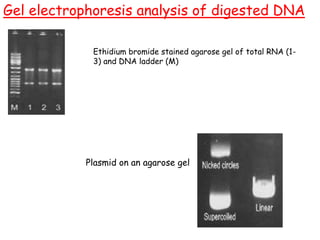
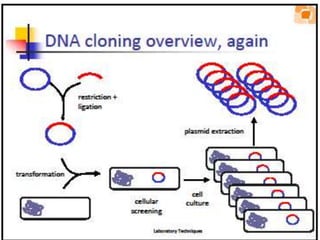
This document provides an overview of basic laboratory techniques in molecular biology, emphasizing safety procedures and essential methodologies. Key techniques discussed include DNA isolation, restriction enzyme usage, and the polymerase chain reaction (PCR), which allows for the selective amplification of DNA. The document serves as a guideline for working with DNA and genetic materials in a laboratory setting.