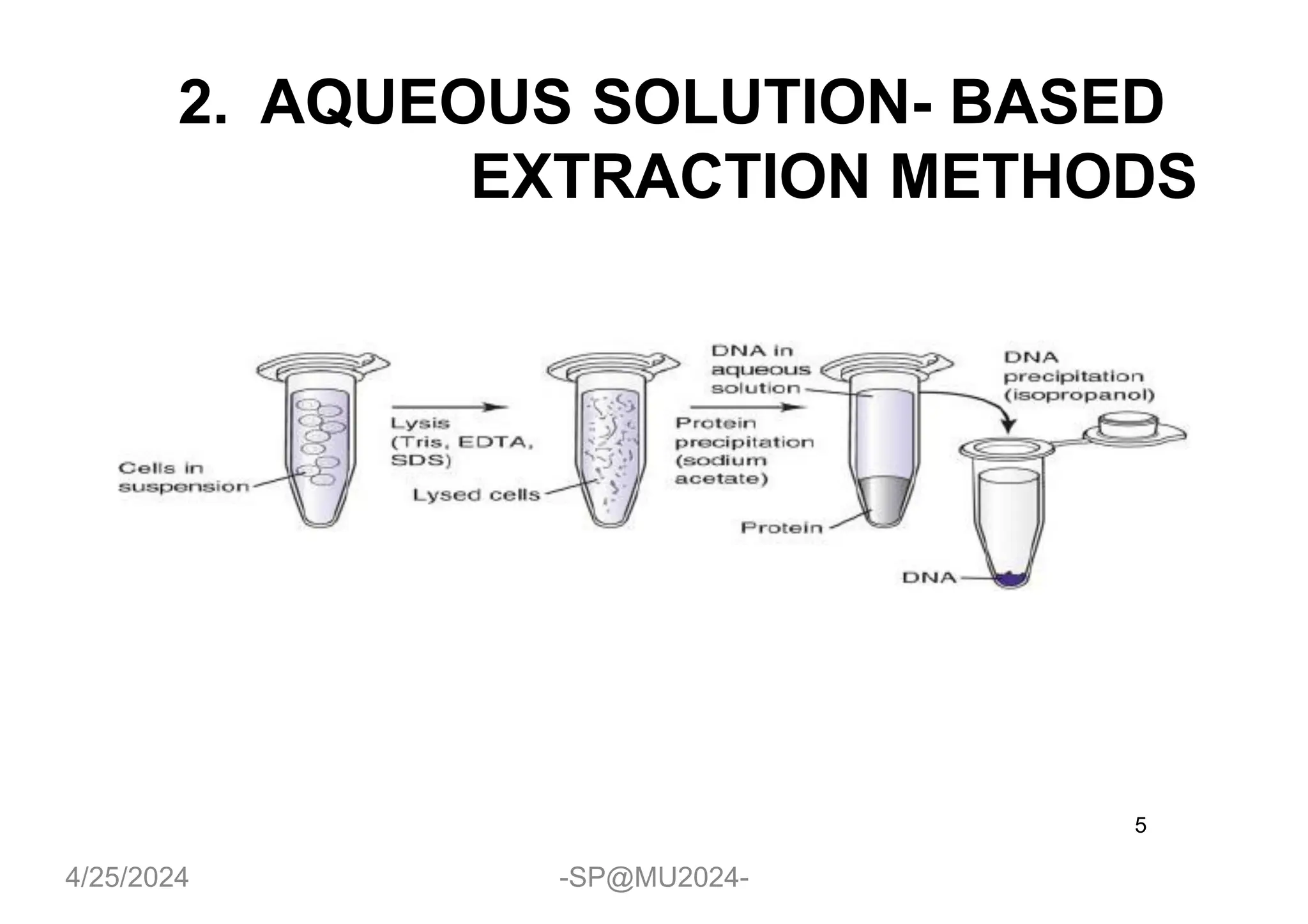
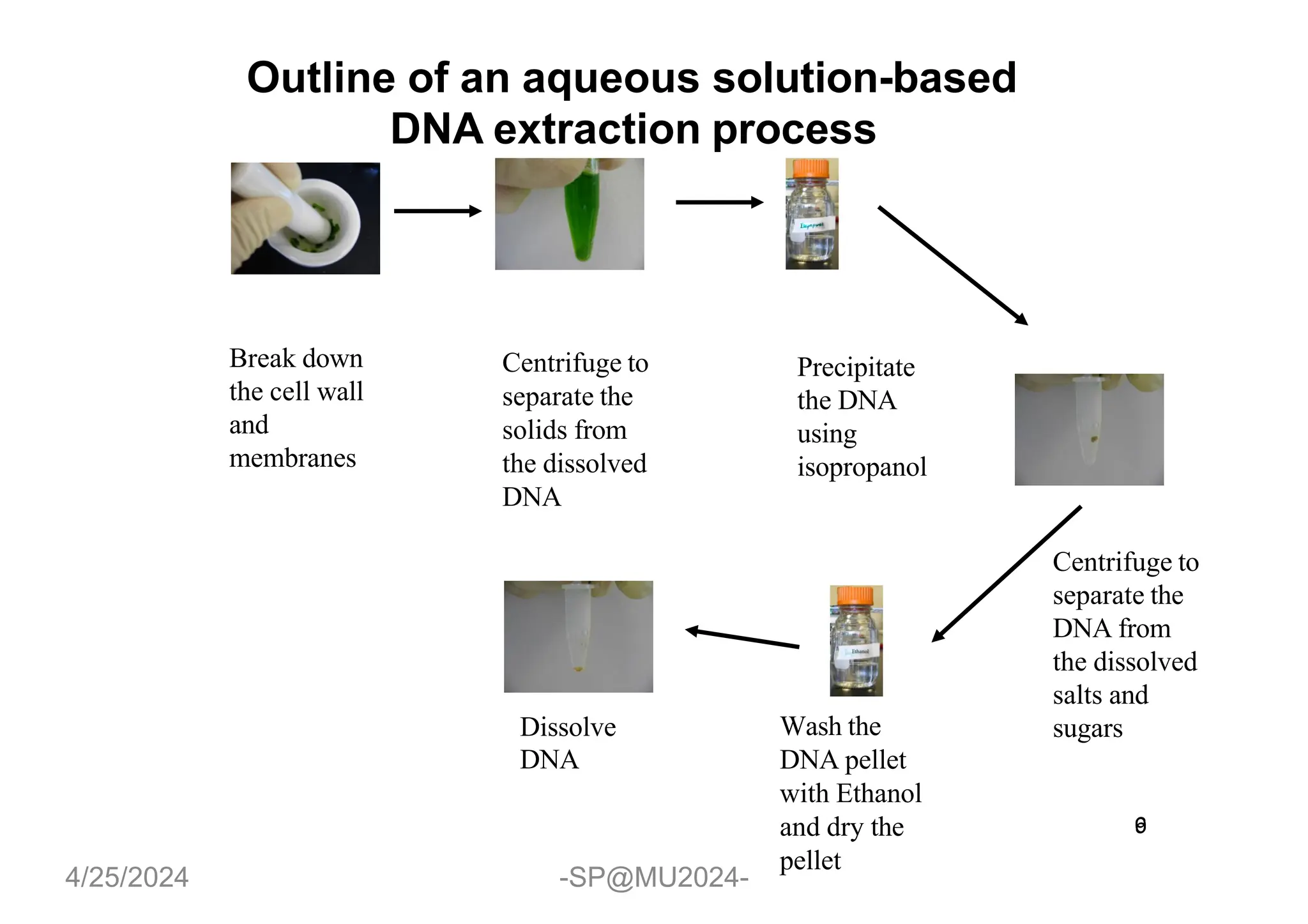
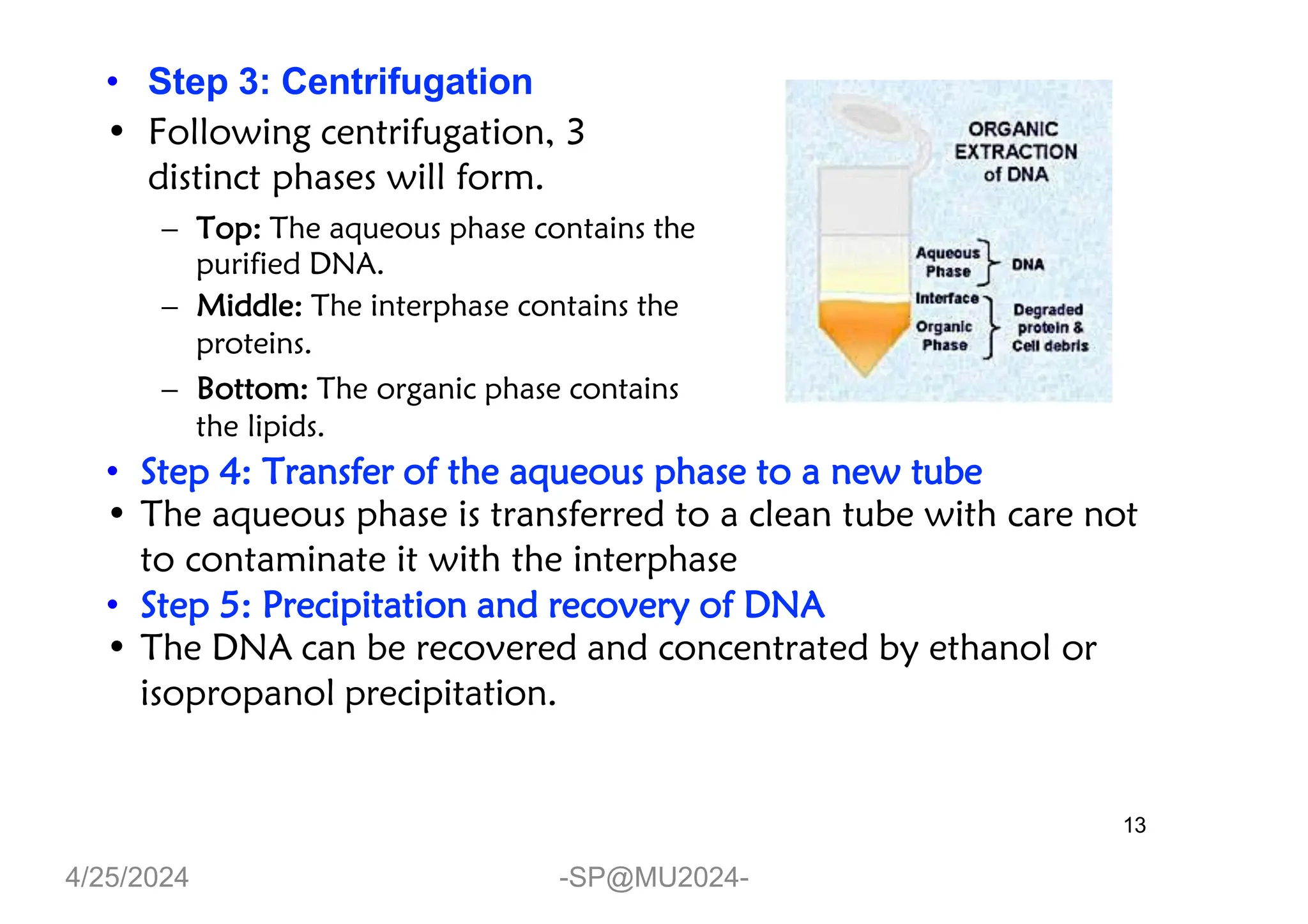
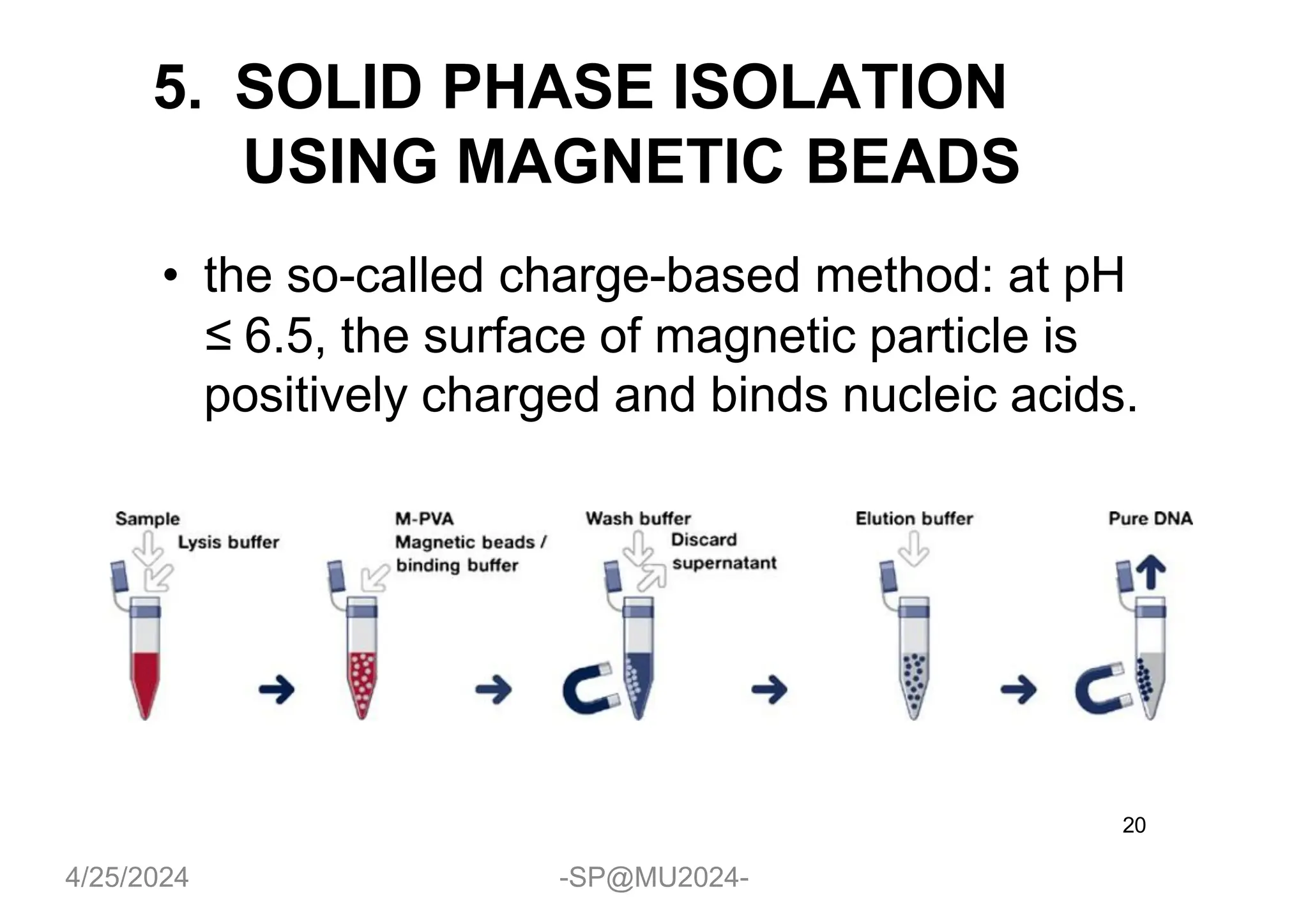
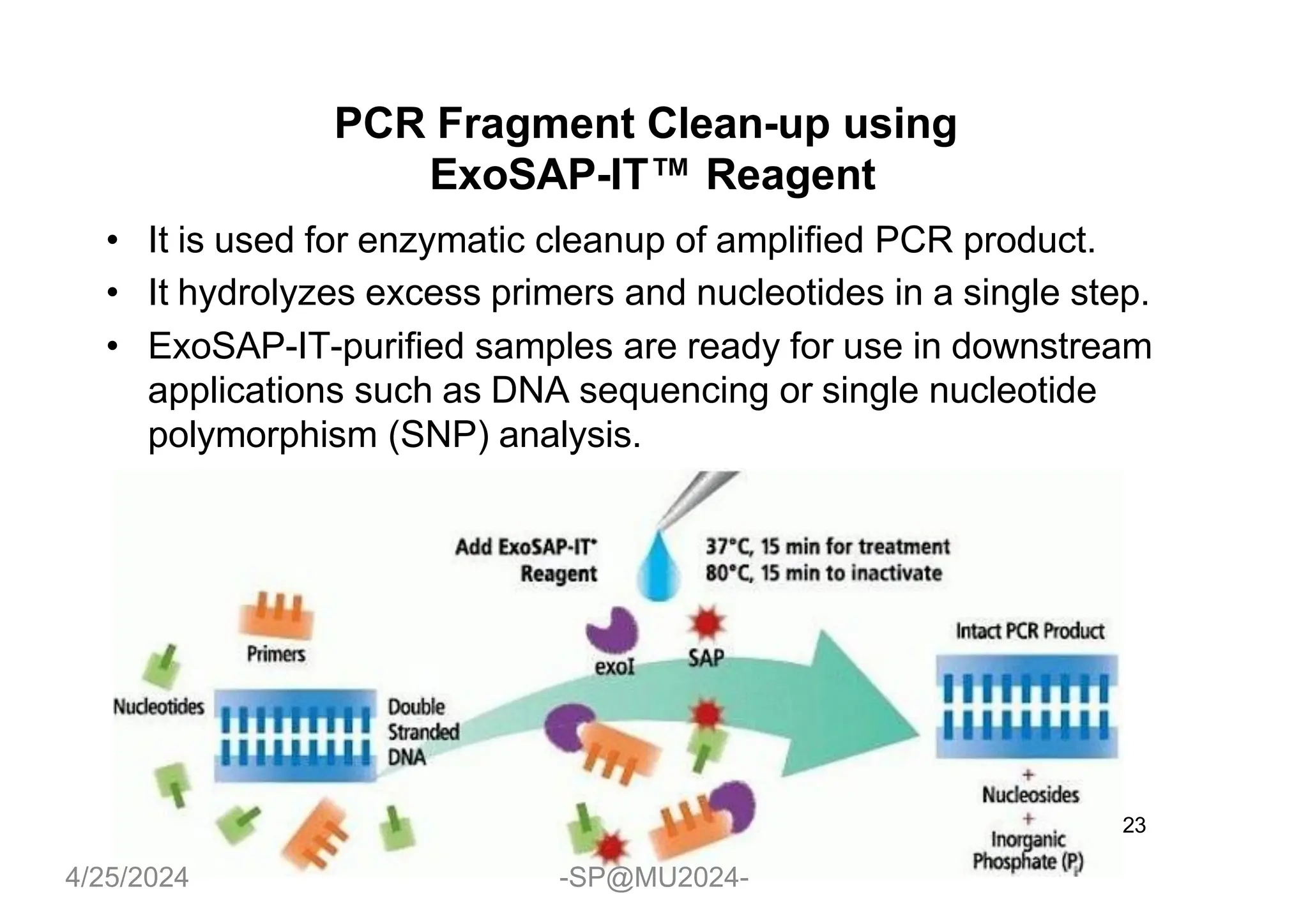
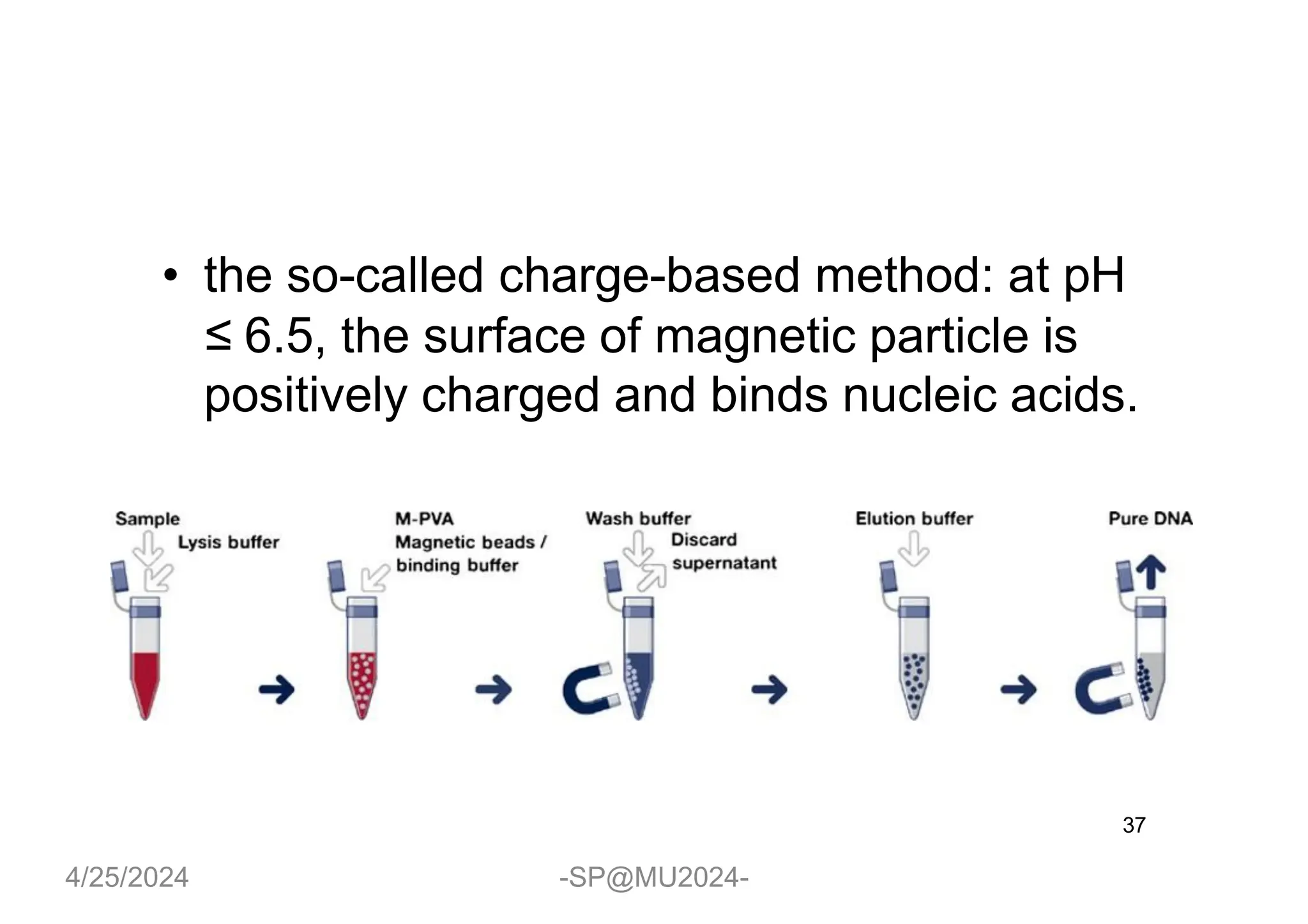
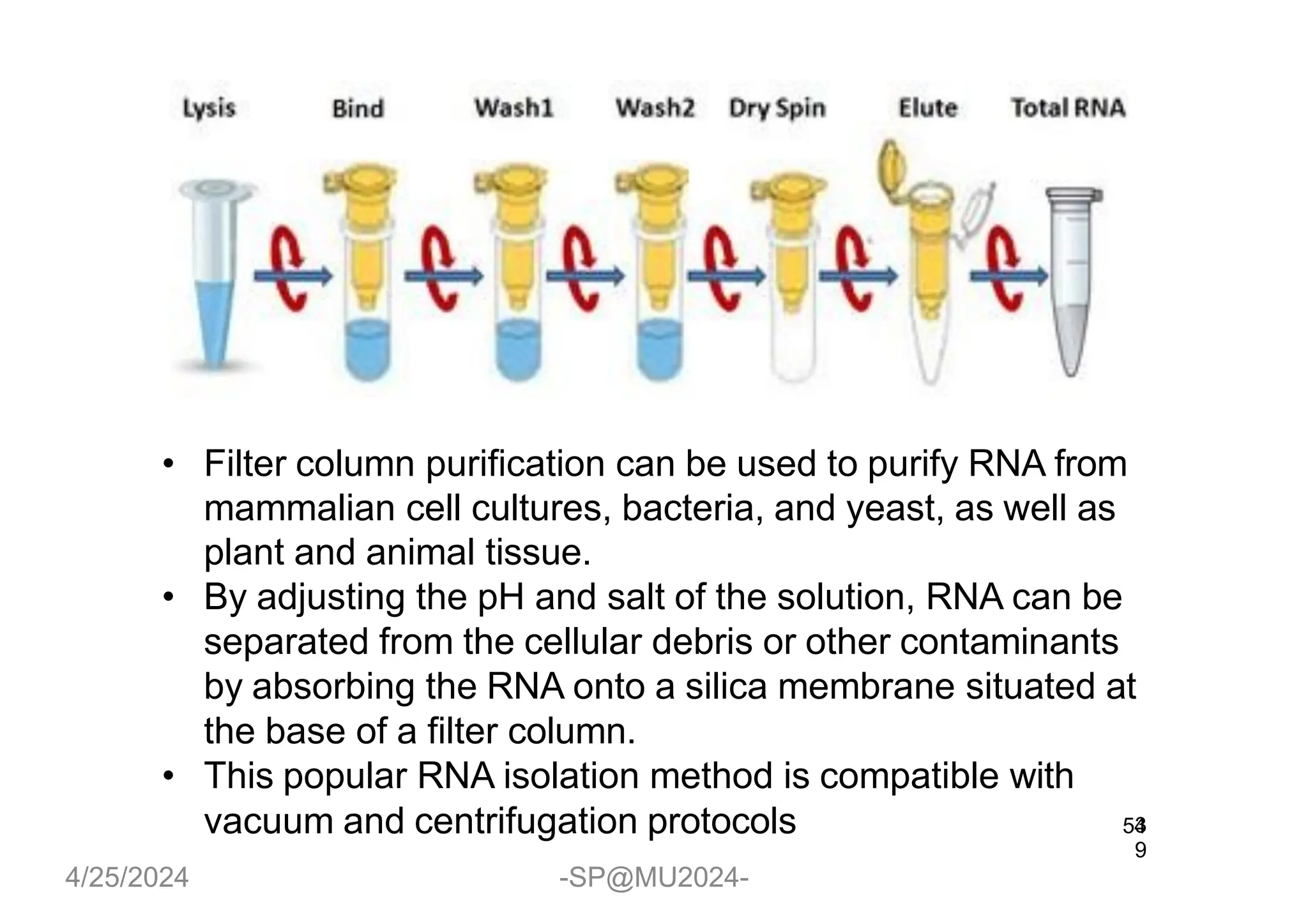
The document outlines methods for DNA and RNA extraction, emphasizing their importance for applications such as PCR and sequencing. It details various techniques including thermal, aqueous, organic solvent-based, and solid phase extraction methods, along with specific procedures for mitochondrial DNA and RNA extraction. Additionally, it covers post-extraction processes such as purification, concentration, and confirming nucleic acid quality for downstream applications.