- The document discusses various approaches for applying machine learning and artificial intelligence to drug discovery.
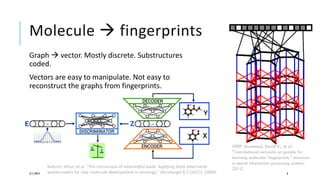
- It describes how molecules and proteins can be represented as graphs, fingerprints, or sequences to be used as input for models.
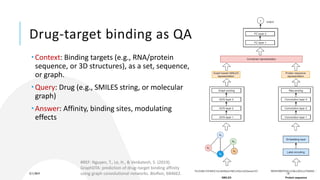
- Different tasks in drug discovery like target binding prediction, generative design of new molecules, and drug repurposing are framed as questions that AI models can aim to answer.
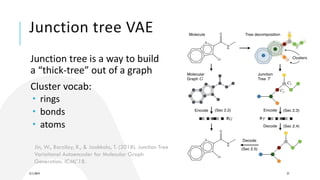
- Techniques discussed include graph neural networks, reinforcement learning, and conditional generation using techniques like translation models.
- Several recent works applying these approaches for tasks like predicting drug-target interactions and generating synthesizable molecules are referenced.