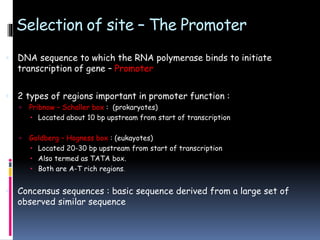
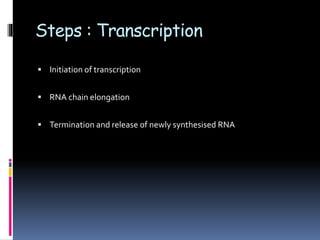
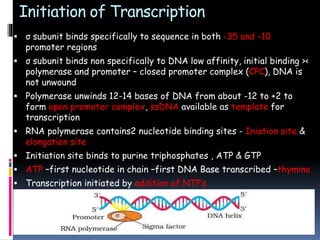
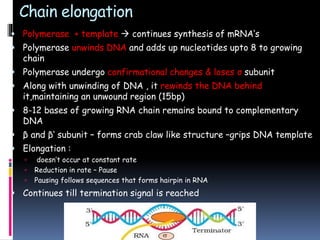
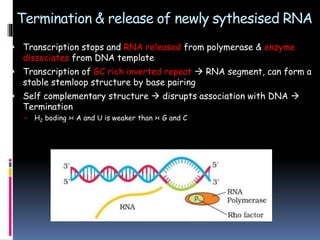
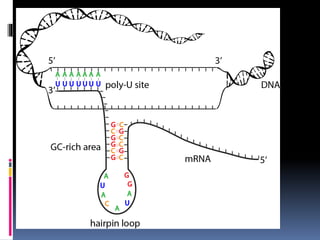
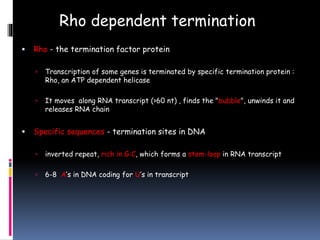
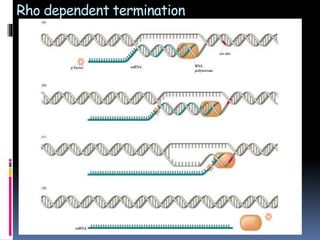
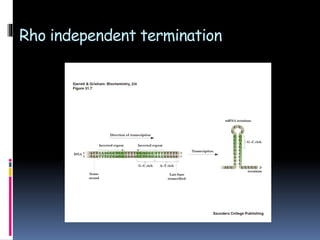
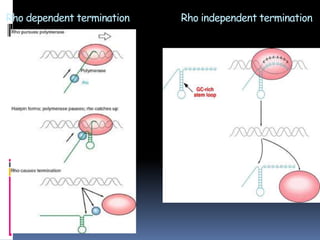
Prokaryotic transcription involves RNA polymerases catalyzing the synthesis of RNA molecules using a DNA template. There are three main steps: initiation, elongation, and termination. Initiation begins with the binding of RNA polymerase to promoter regions on the DNA. Elongation then occurs as the polymerase unwinds the DNA and adds nucleotides to the growing RNA chain. Termination happens when either Rho-dependent or Rho-independent termination signals are reached, causing the release of the completed RNA molecule.

![ RNA POLYEMERASE:
Complex enzyme – made up of multiple polypeptide chain
6 subunits :
2α subunit – binding of enzyme to promoter region
β – nucleotide addition
β‘ – part of core enzyme , keeps the RNA bound to DNA template
ω – part of enzyme, but no specific function
σ – sigma factor, recognize promoter region binds to it. weakly
bound, which dissociates from enzyme during elongation
[ Core enzyme : σ free unit , ie., α2ββ‘ Holoenzyme : the complete enzyme,
ie., α2ββ‘σ ]
One of the largest enzyme (mw – 465,000Da) ,contact with many
DNA bases simultaneously](https://image.slidesharecdn.com/unit7proktranscription-211207032223/85/Unit-7-prok-transcription-5-320.jpg)