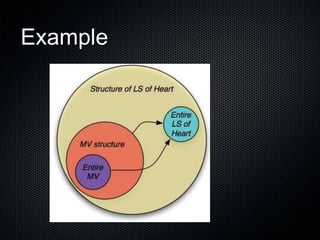
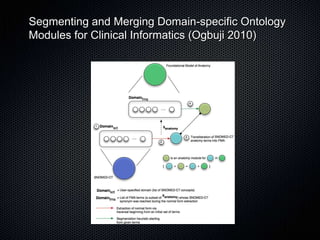
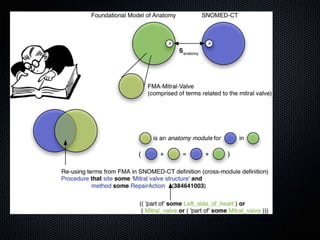
The document discusses the automation of clinical ontology extraction, addressing the need for reproducible methods to fill gaps in medical terminologies and manage the complexities introduced by proliferating domain-specific ontologies. It highlights goals such as customizing large ontologies like SNOMED-CT and developing manageable terminology modules, while utilizing foundational models like the Foundational Model of Anatomy (FMA) to preserve definitions and support quality assurance. Additionally, the document introduces techniques for segmenting and merging ontologies to enhance clinical informatics applications.