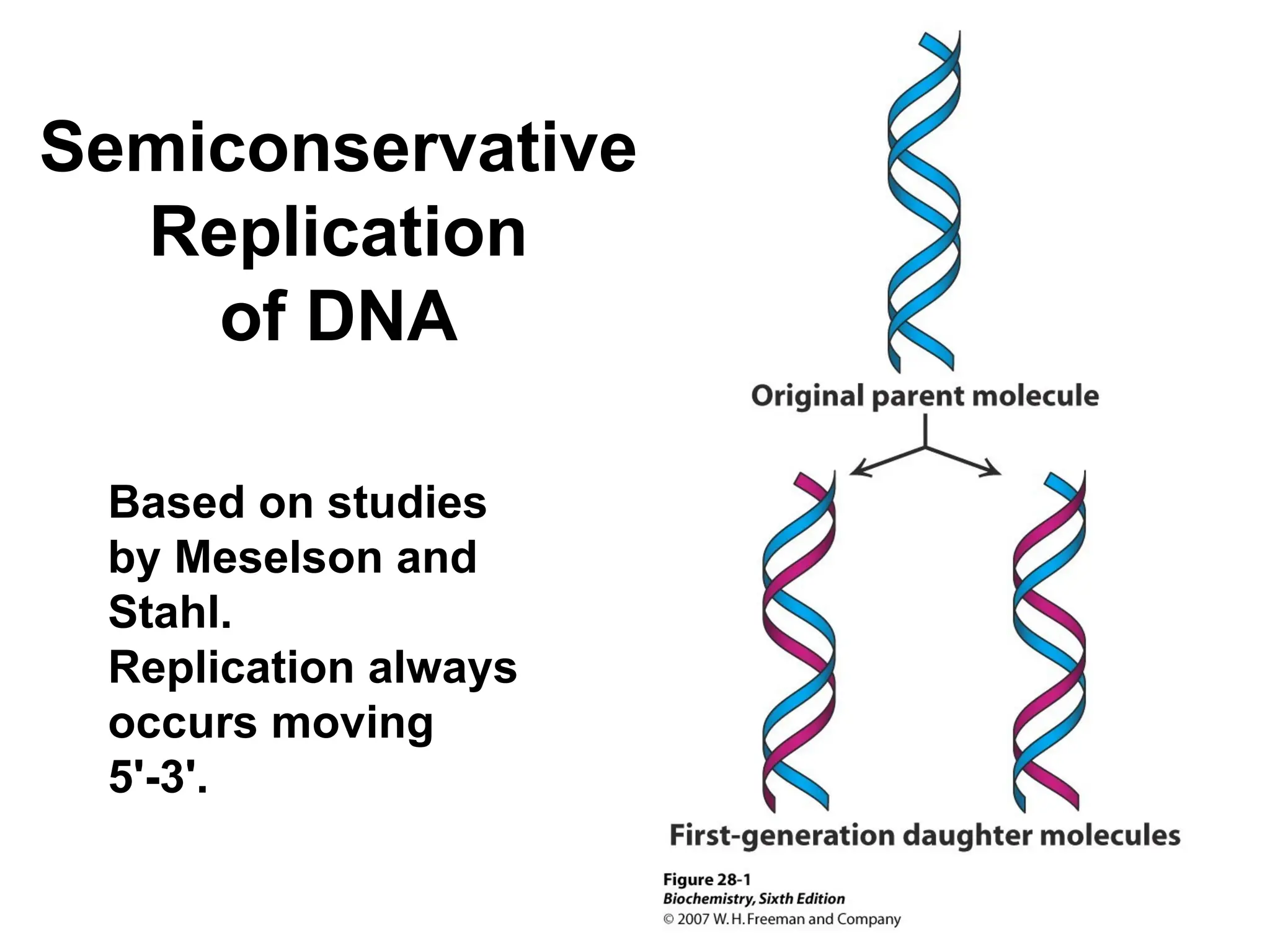
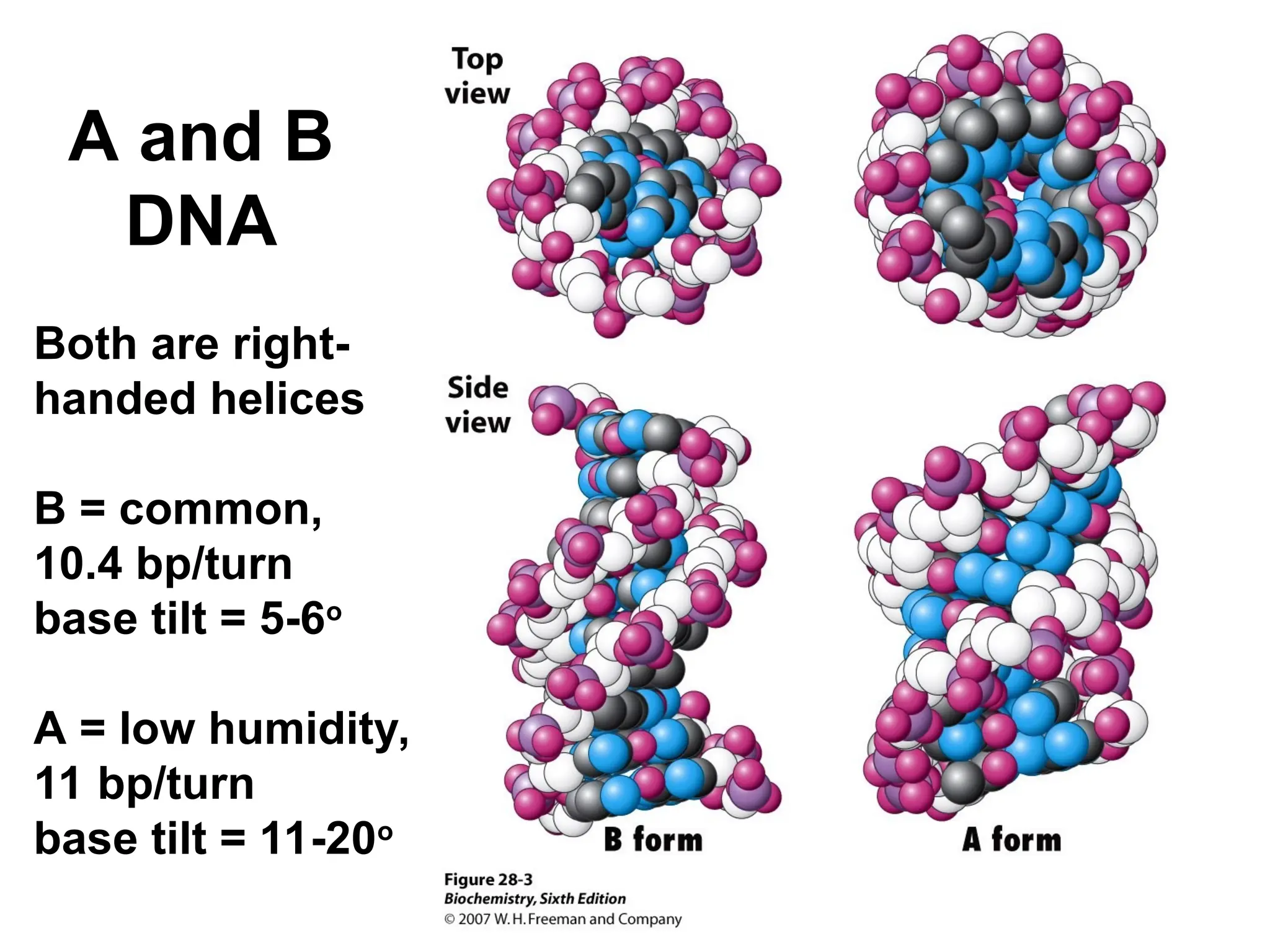
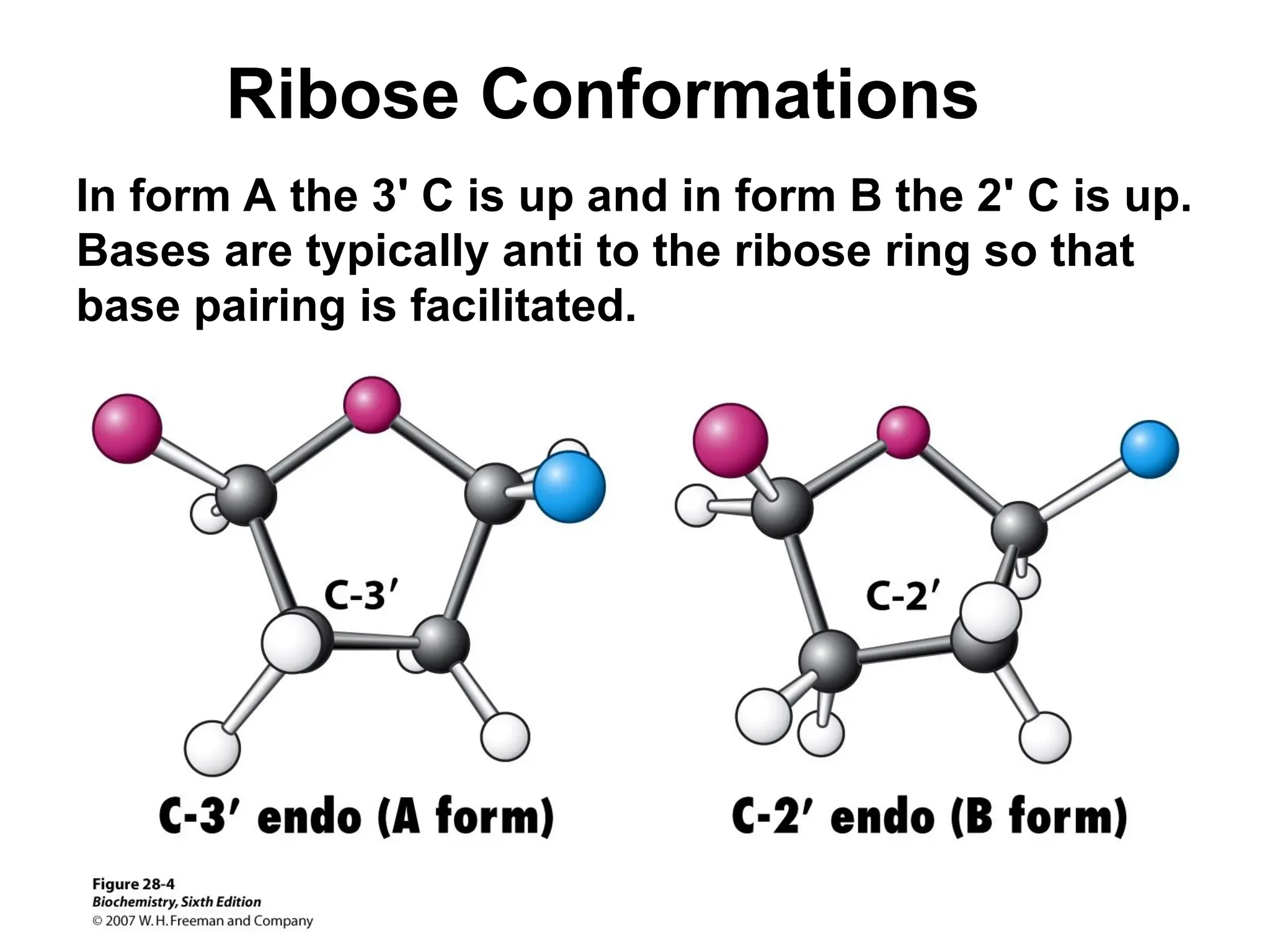
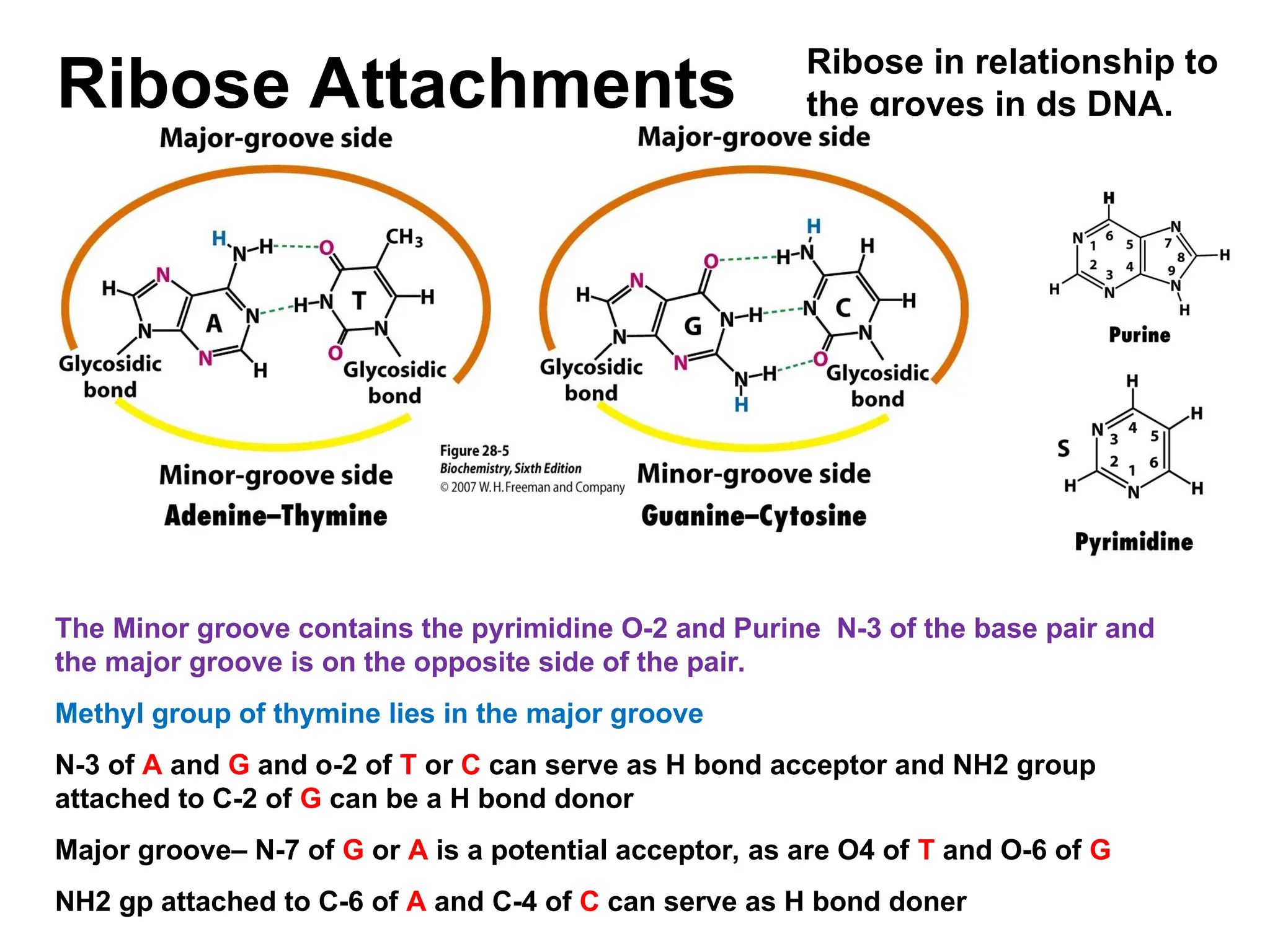
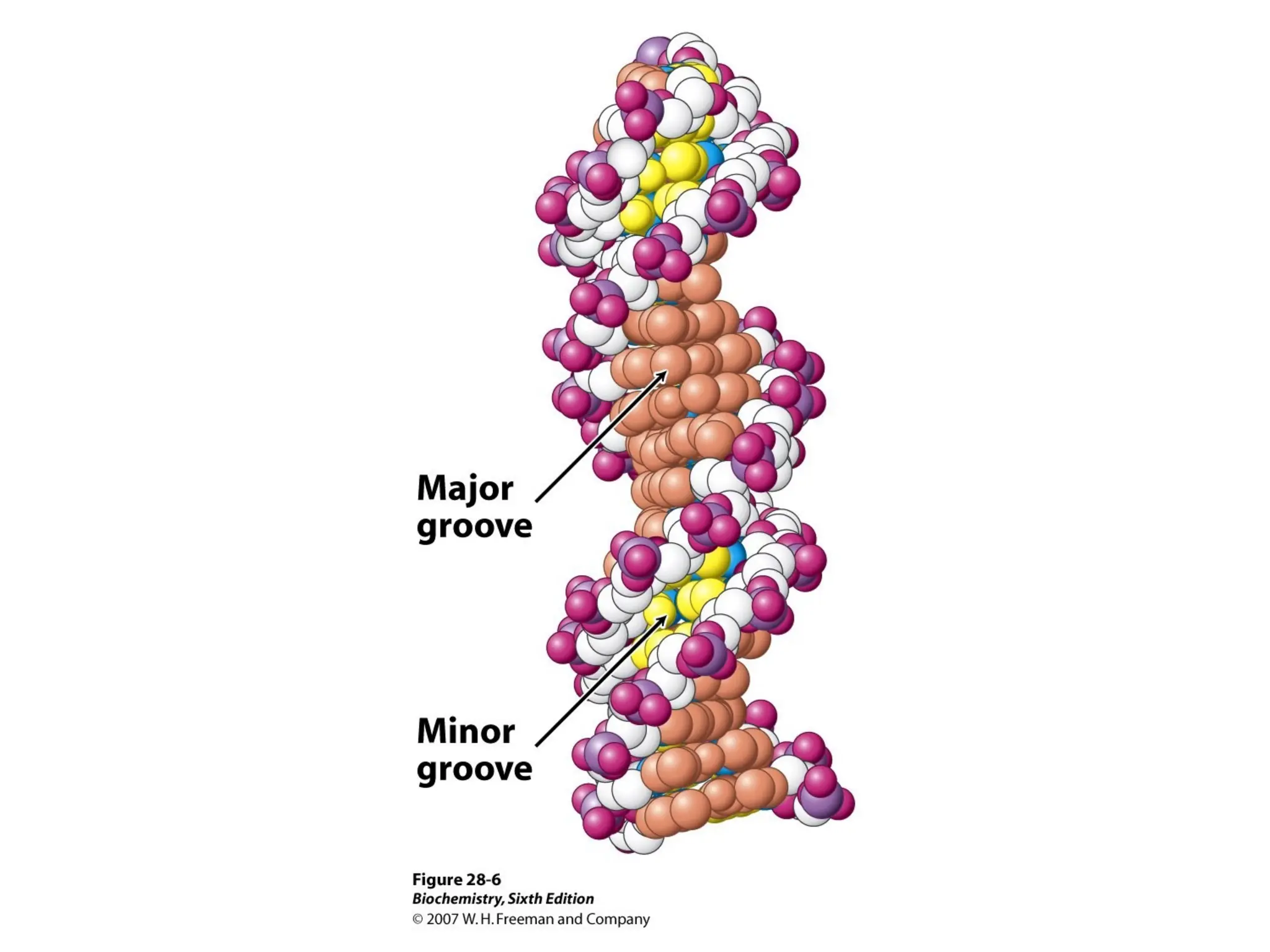
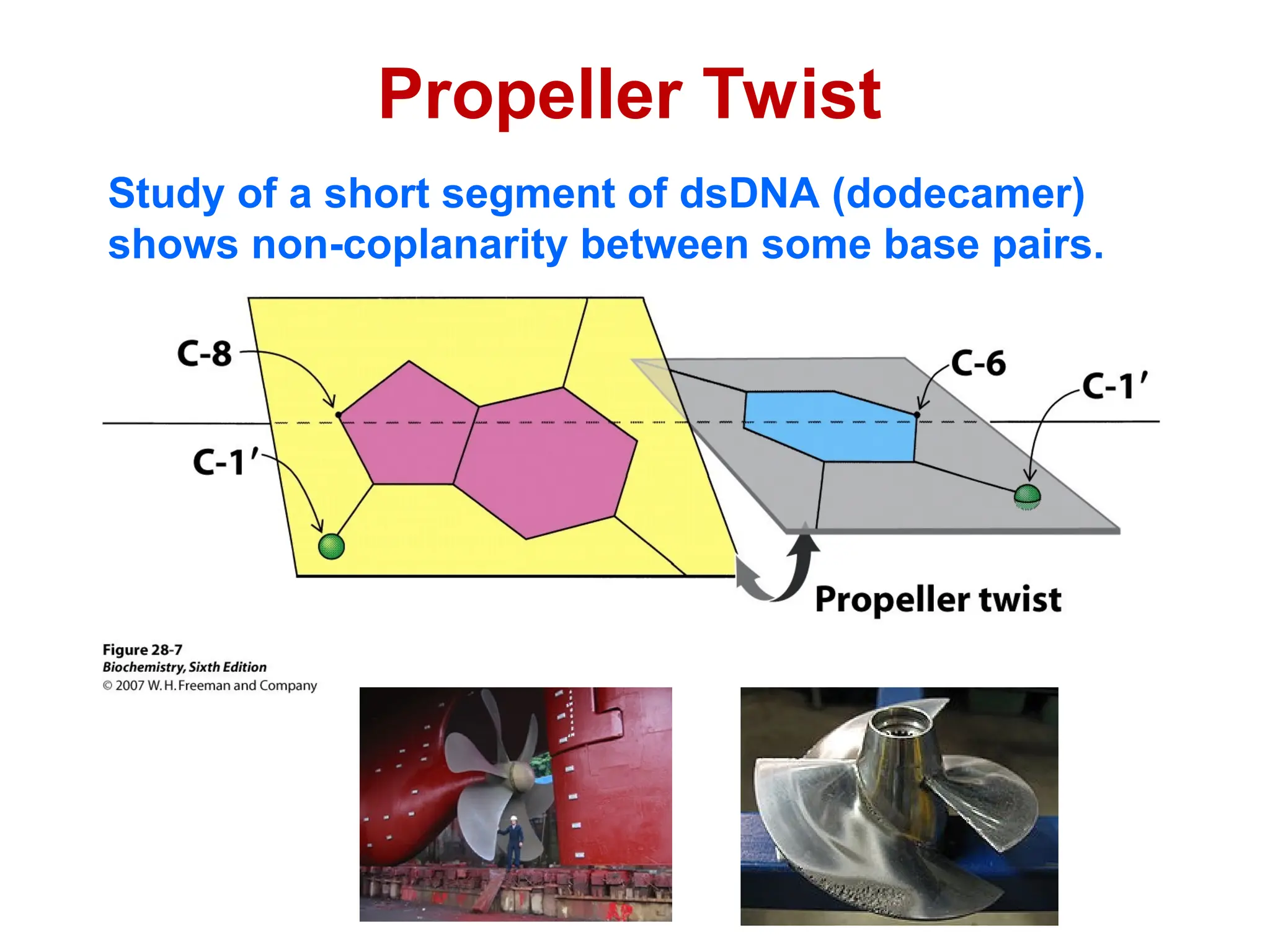
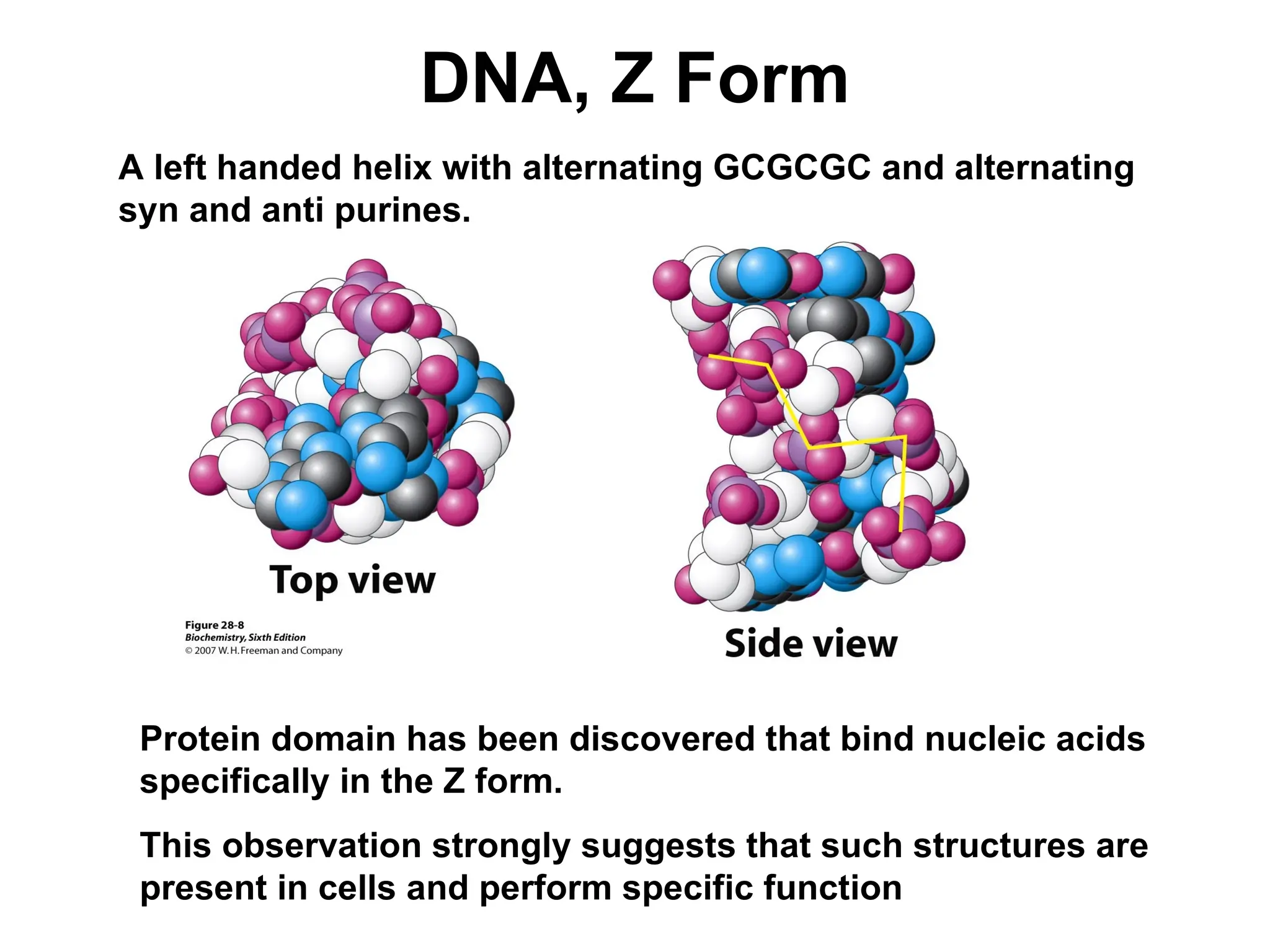
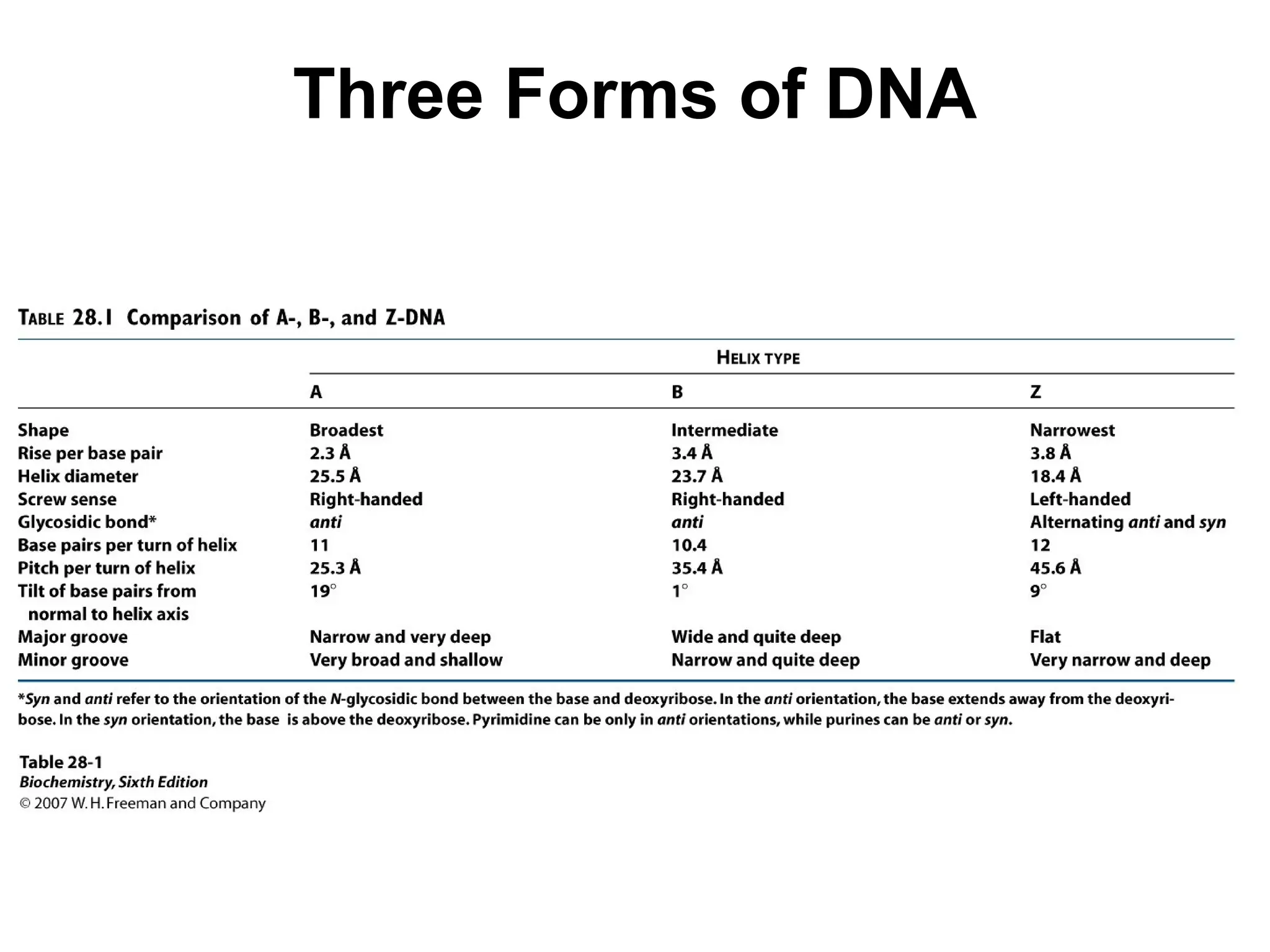
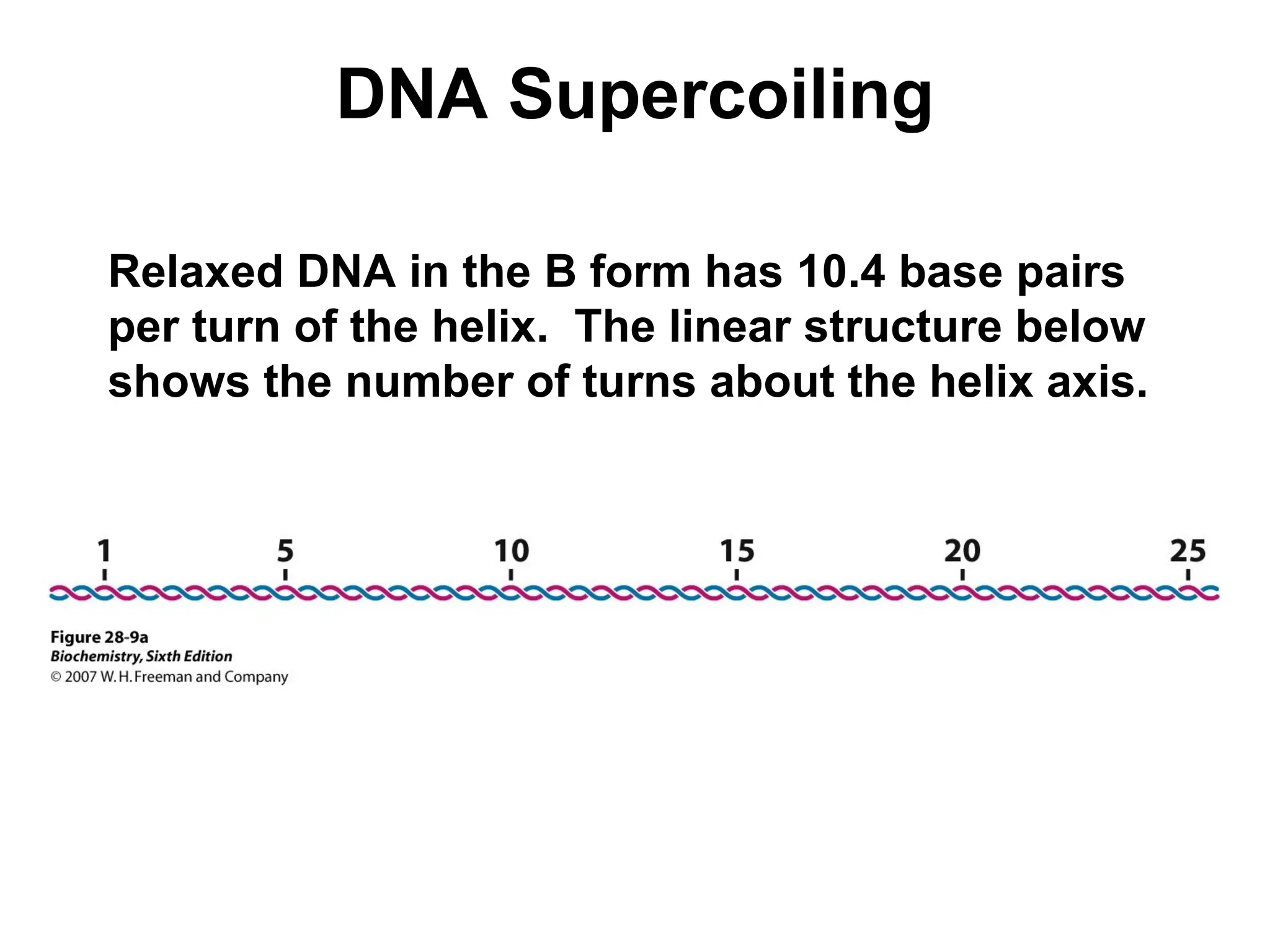
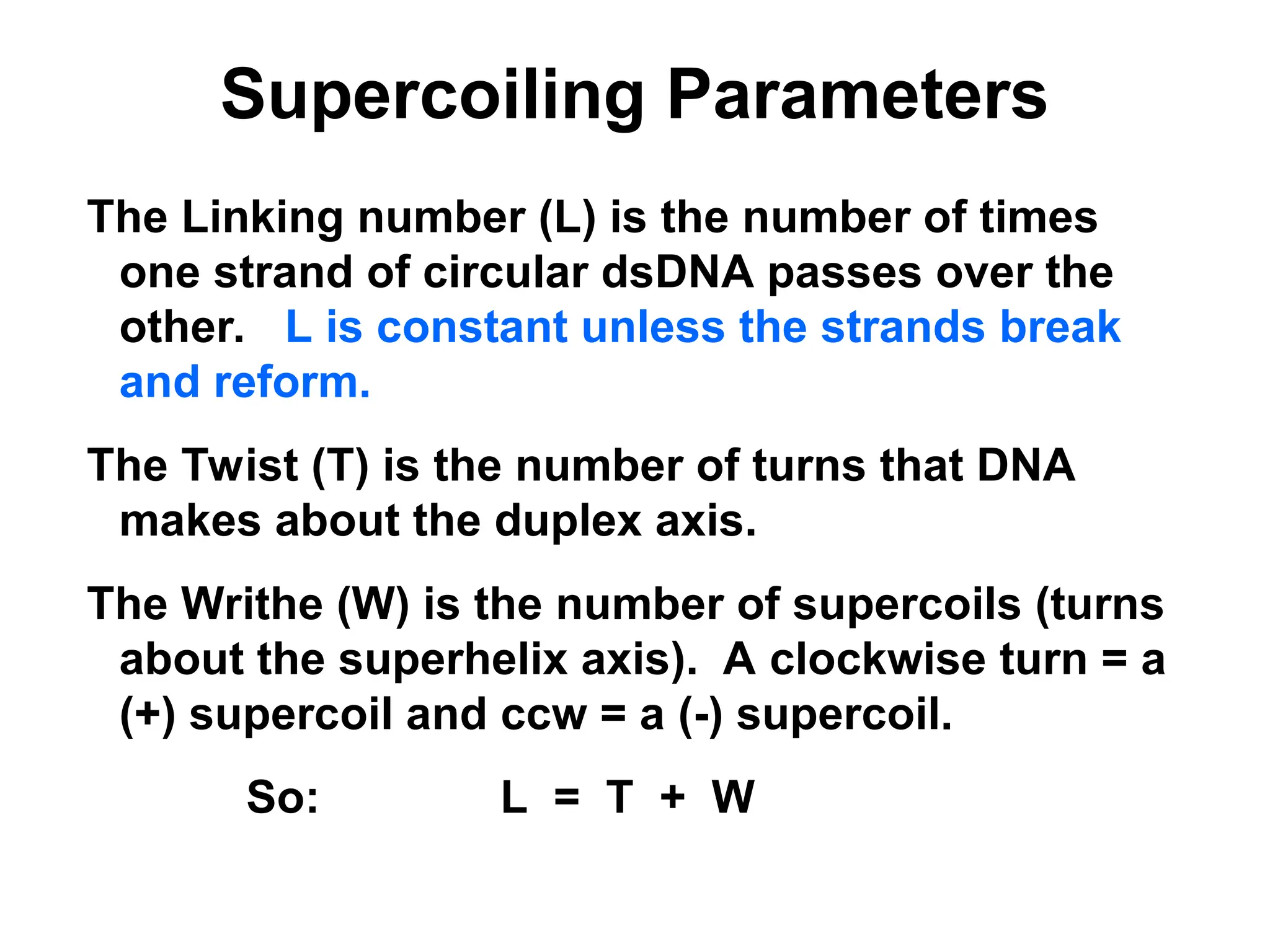
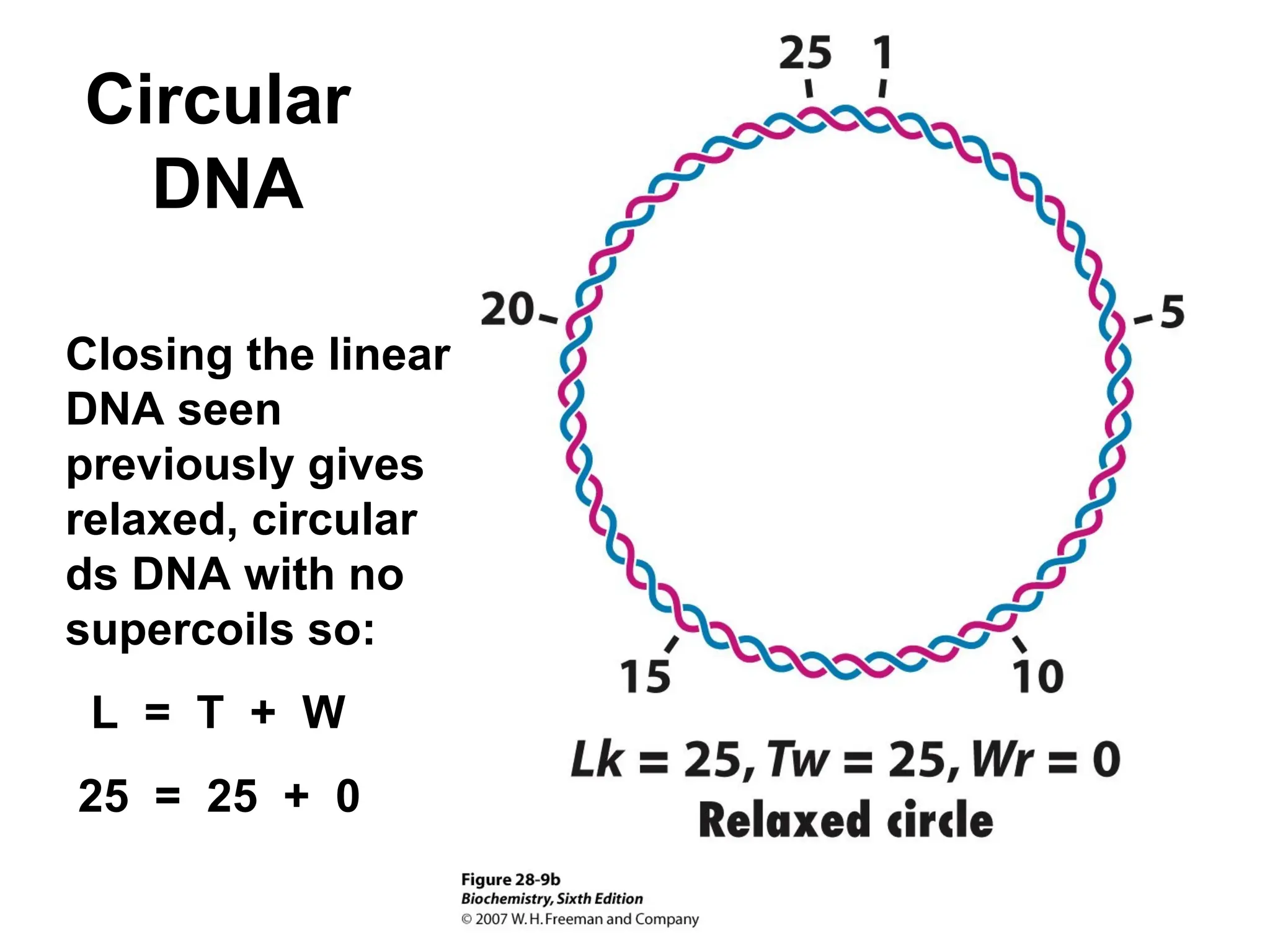
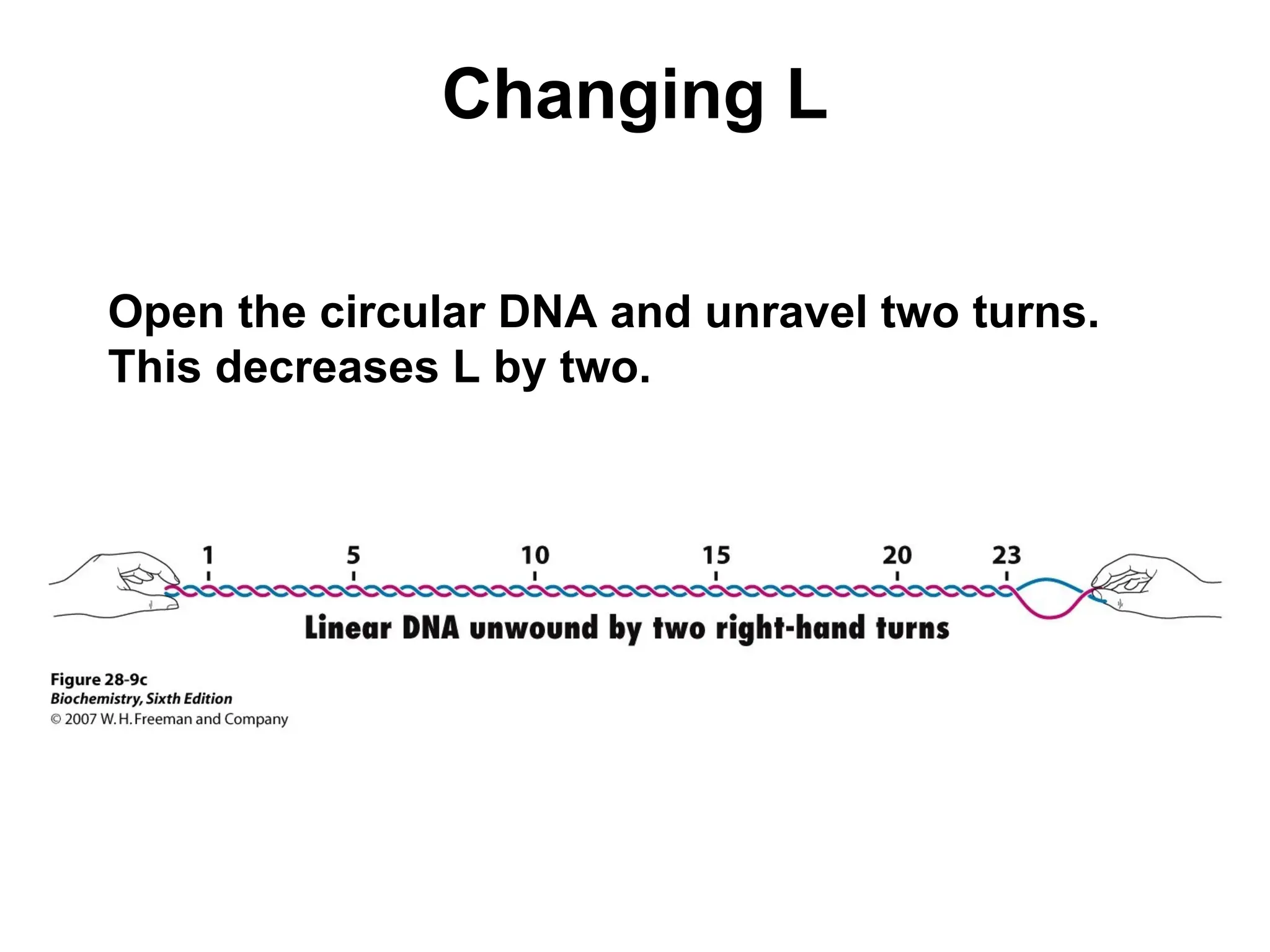
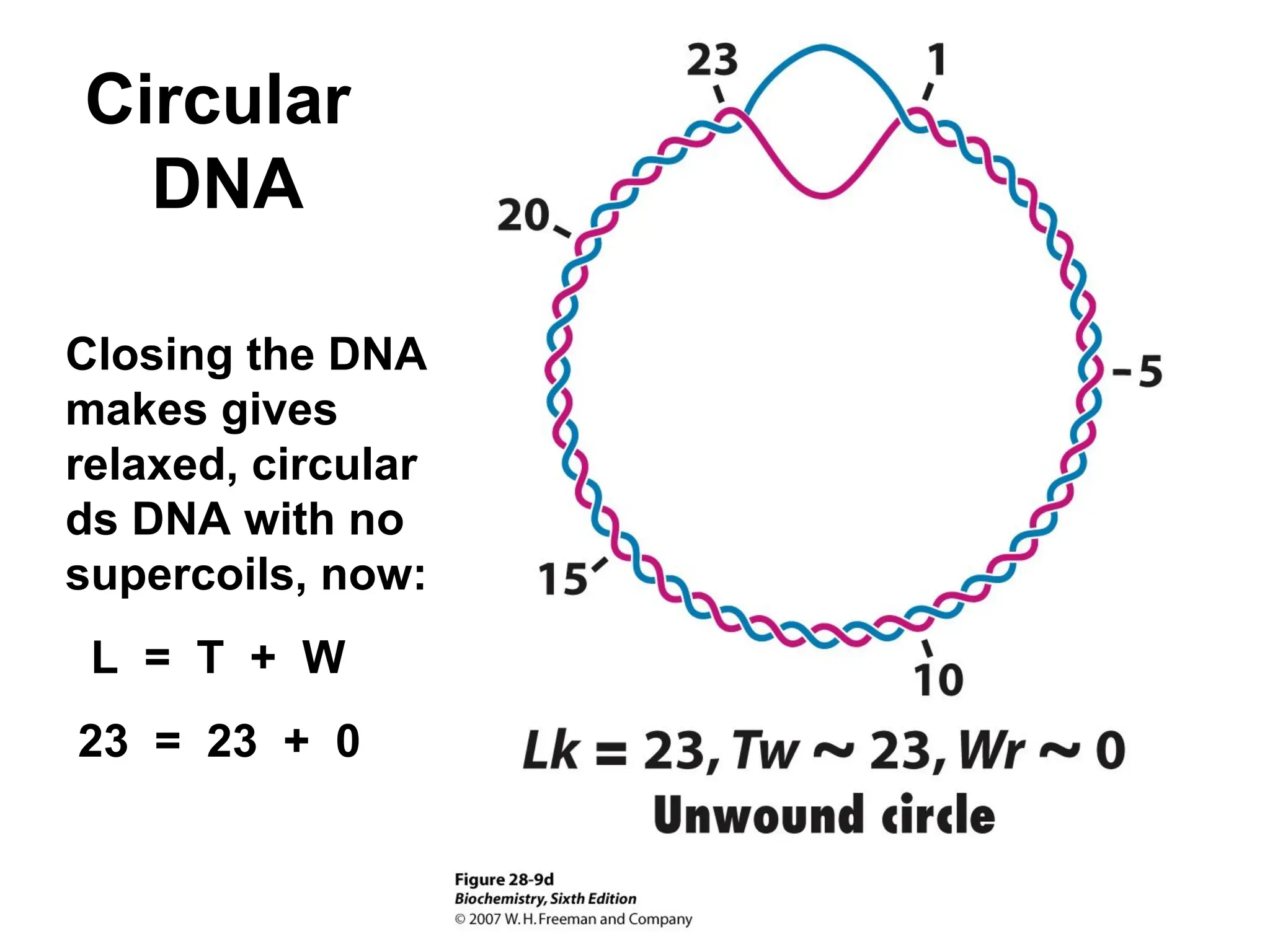
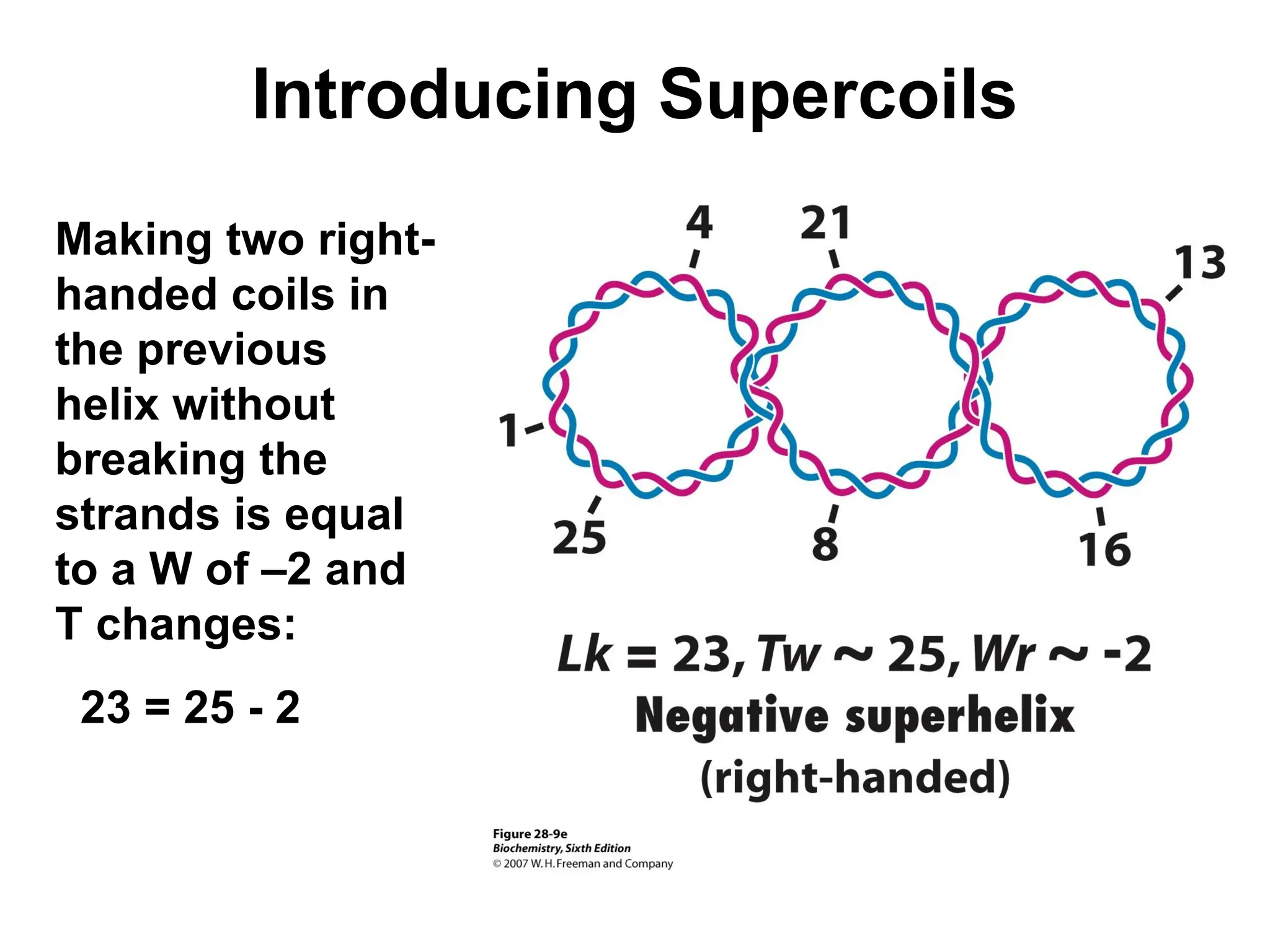
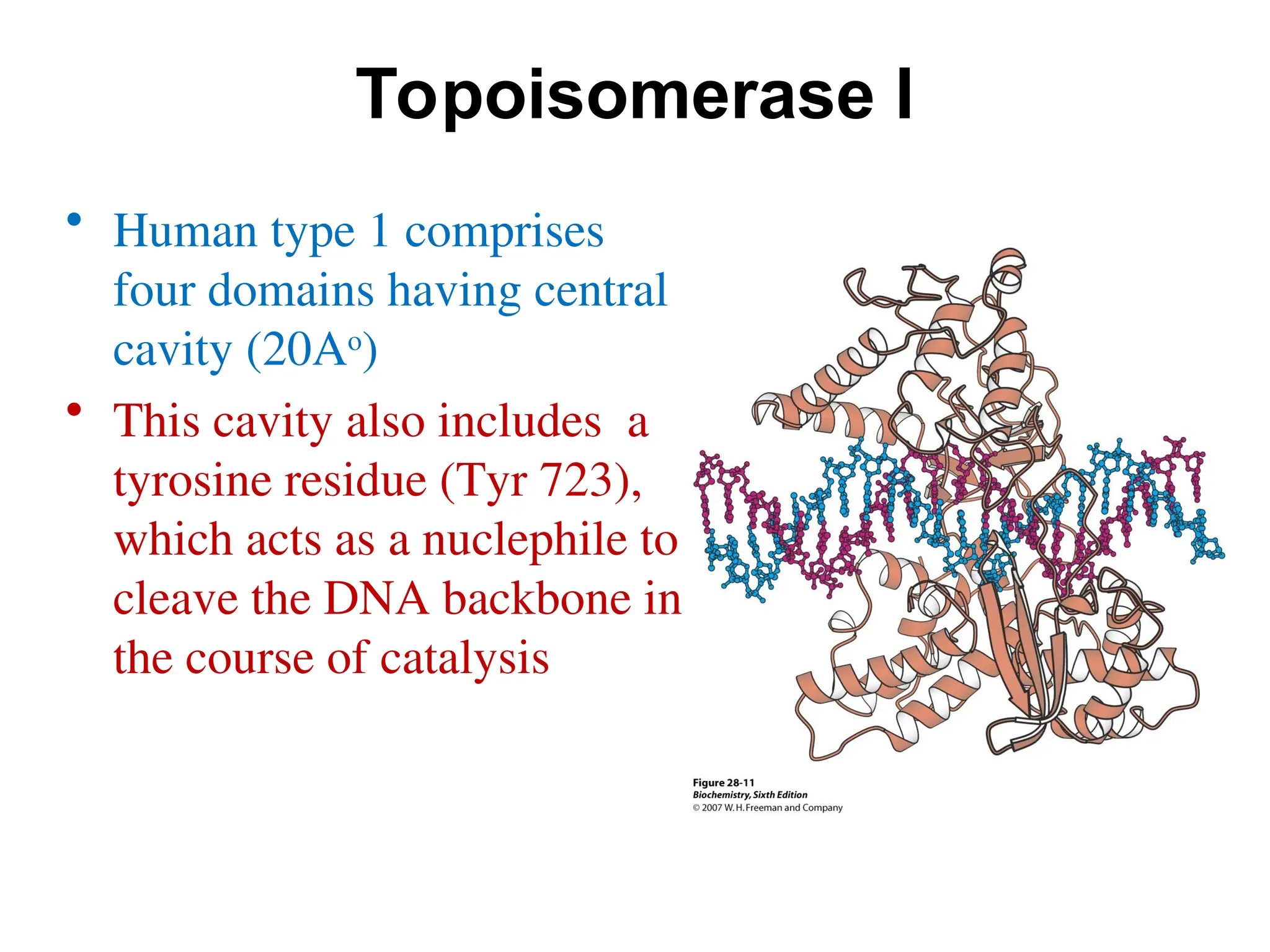
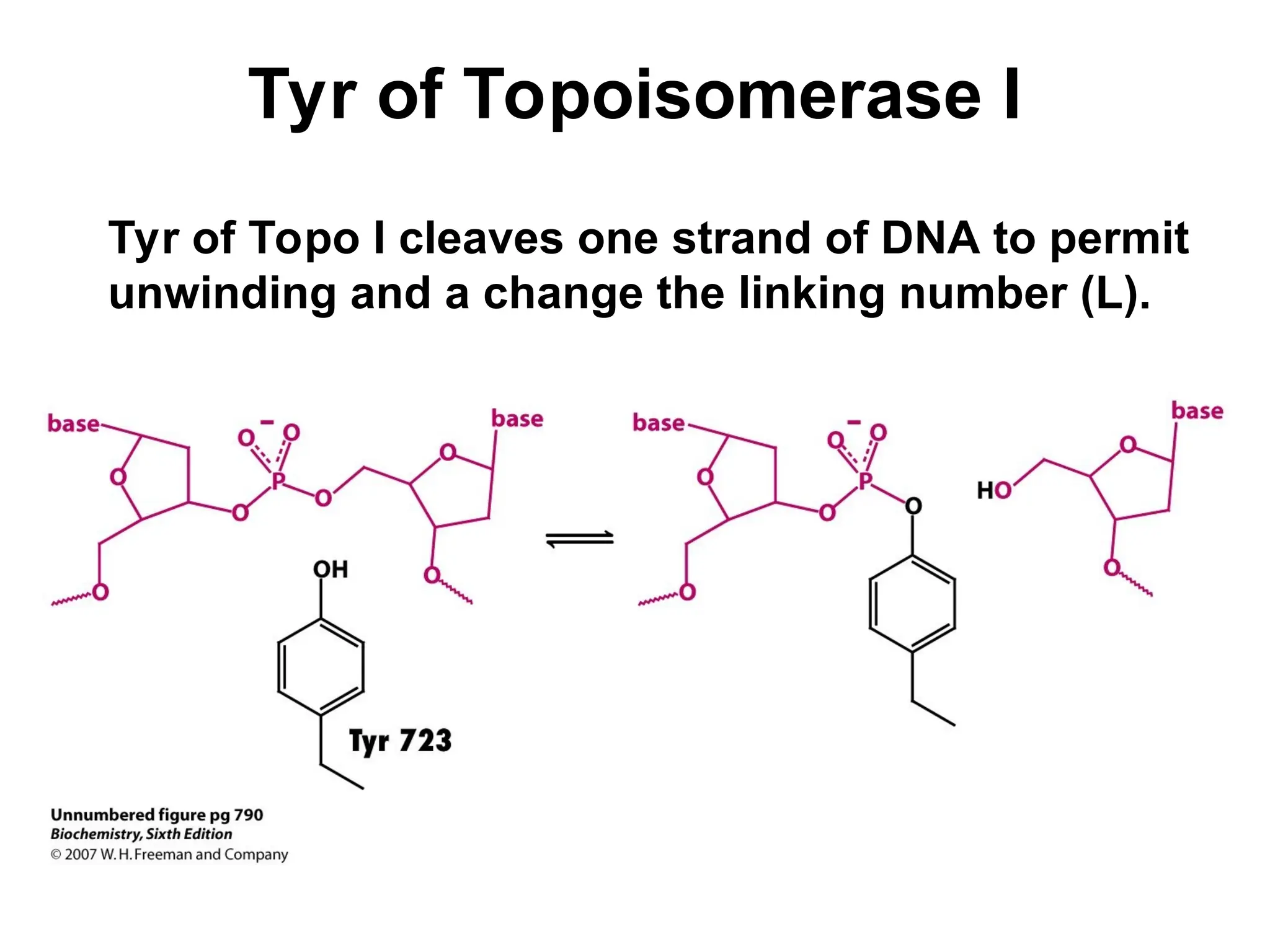
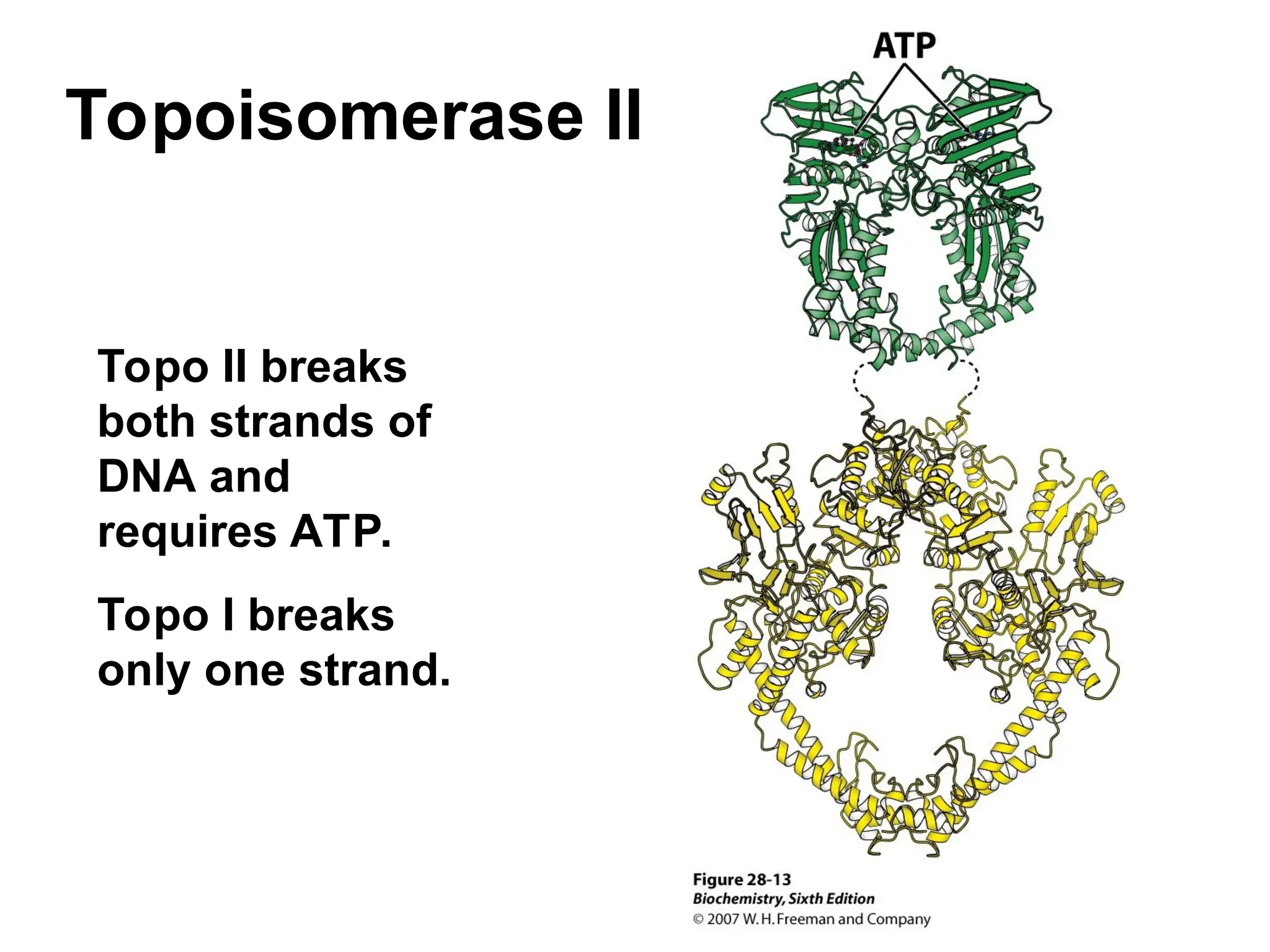
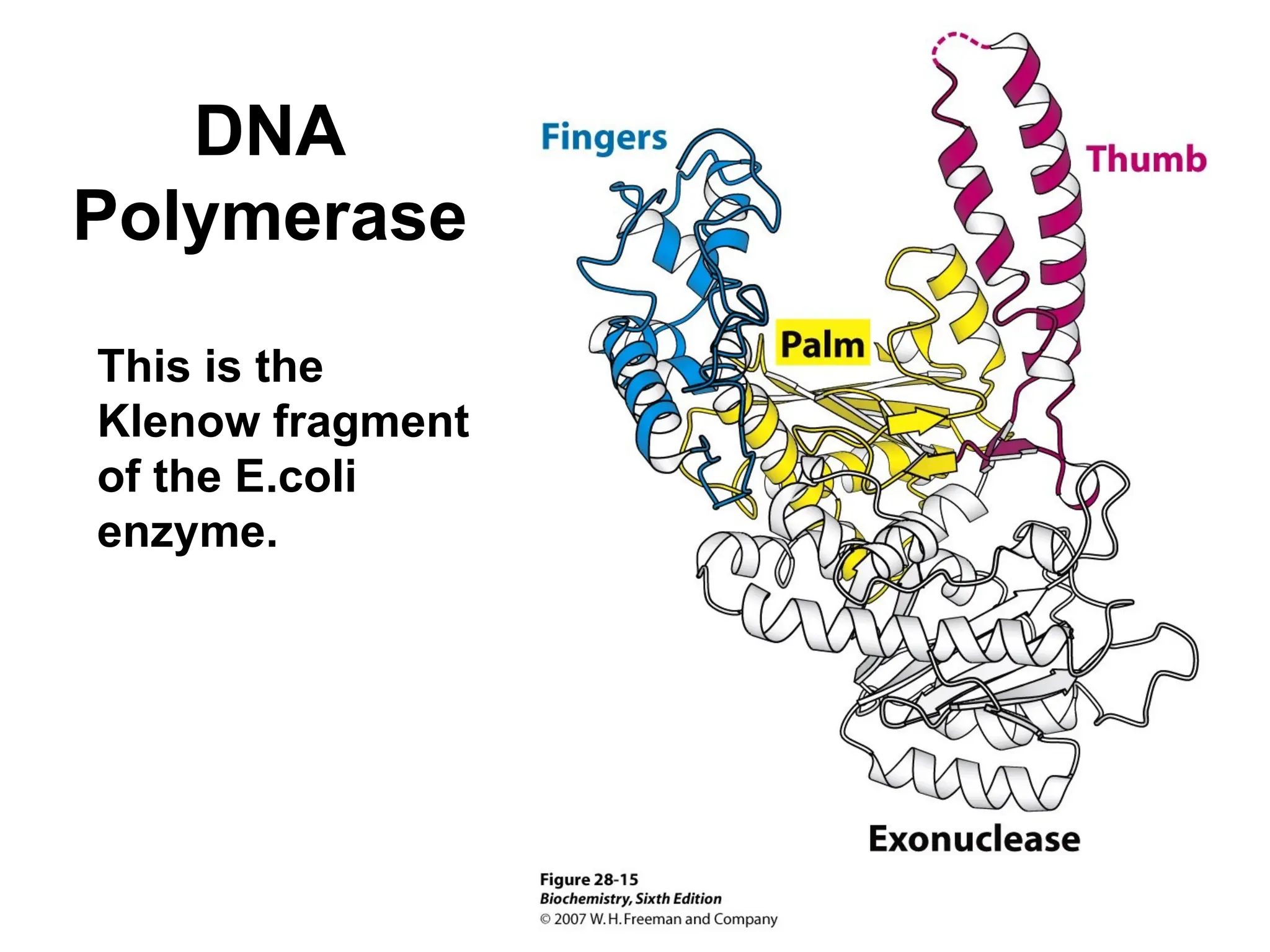
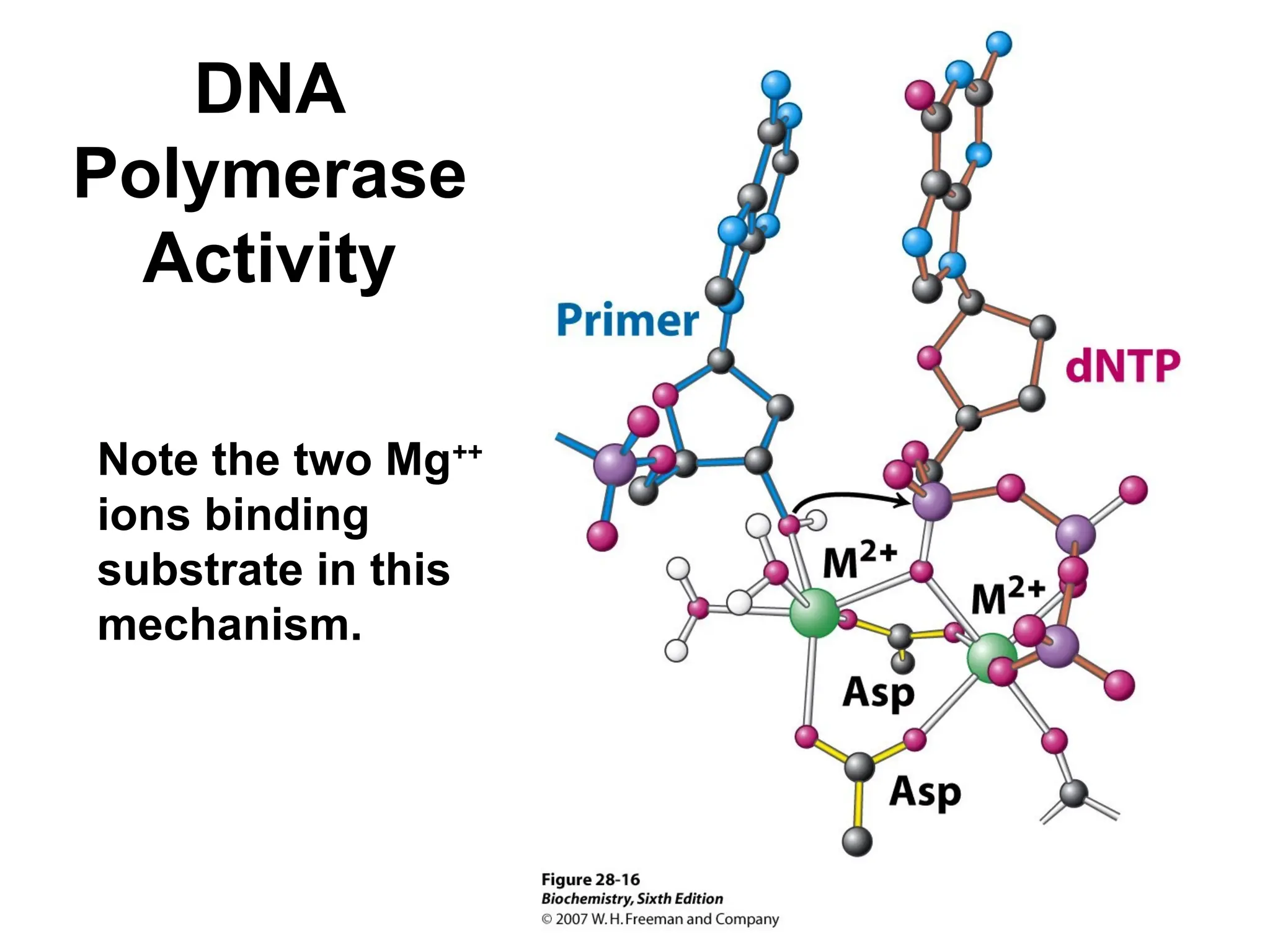
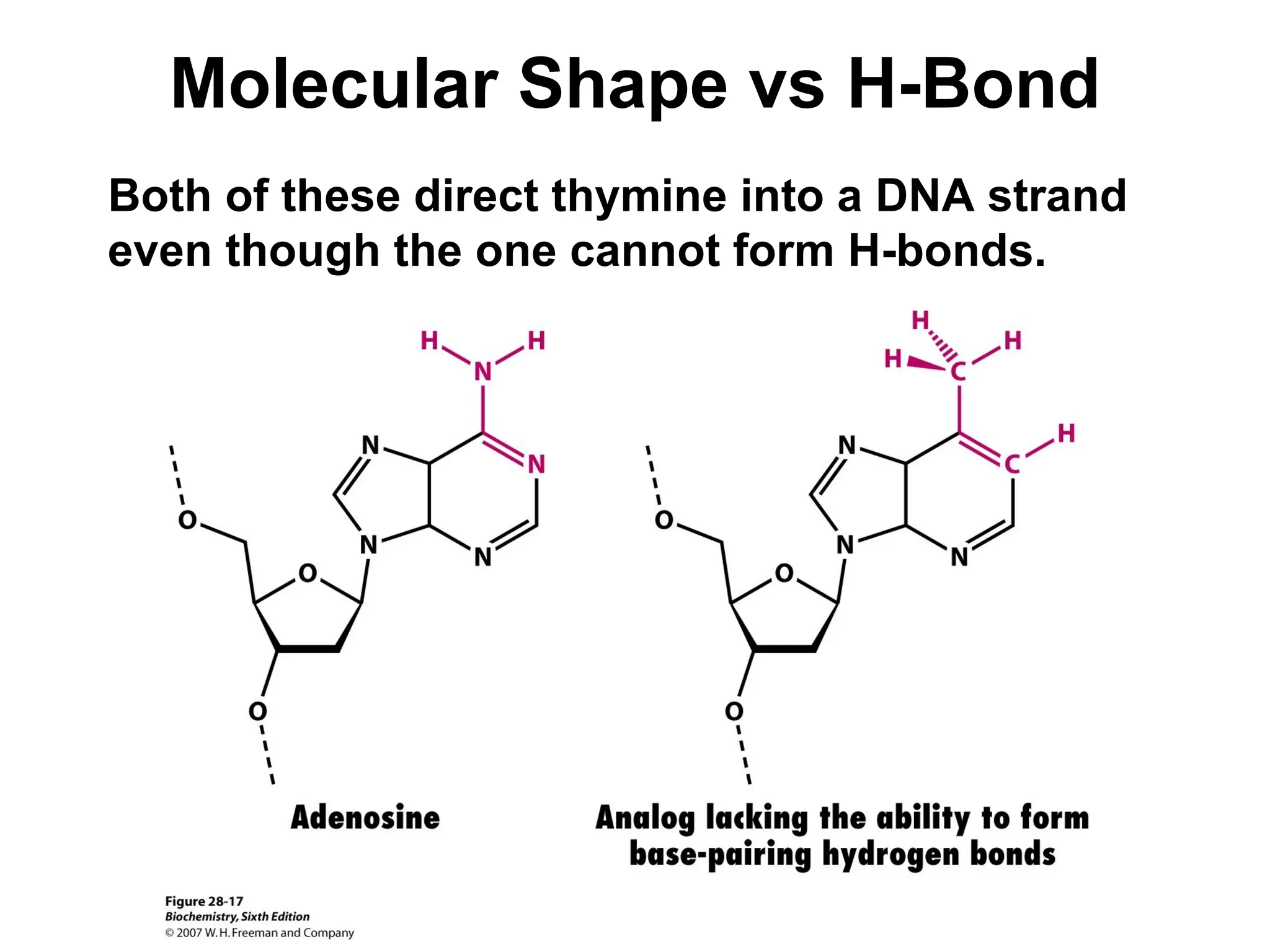
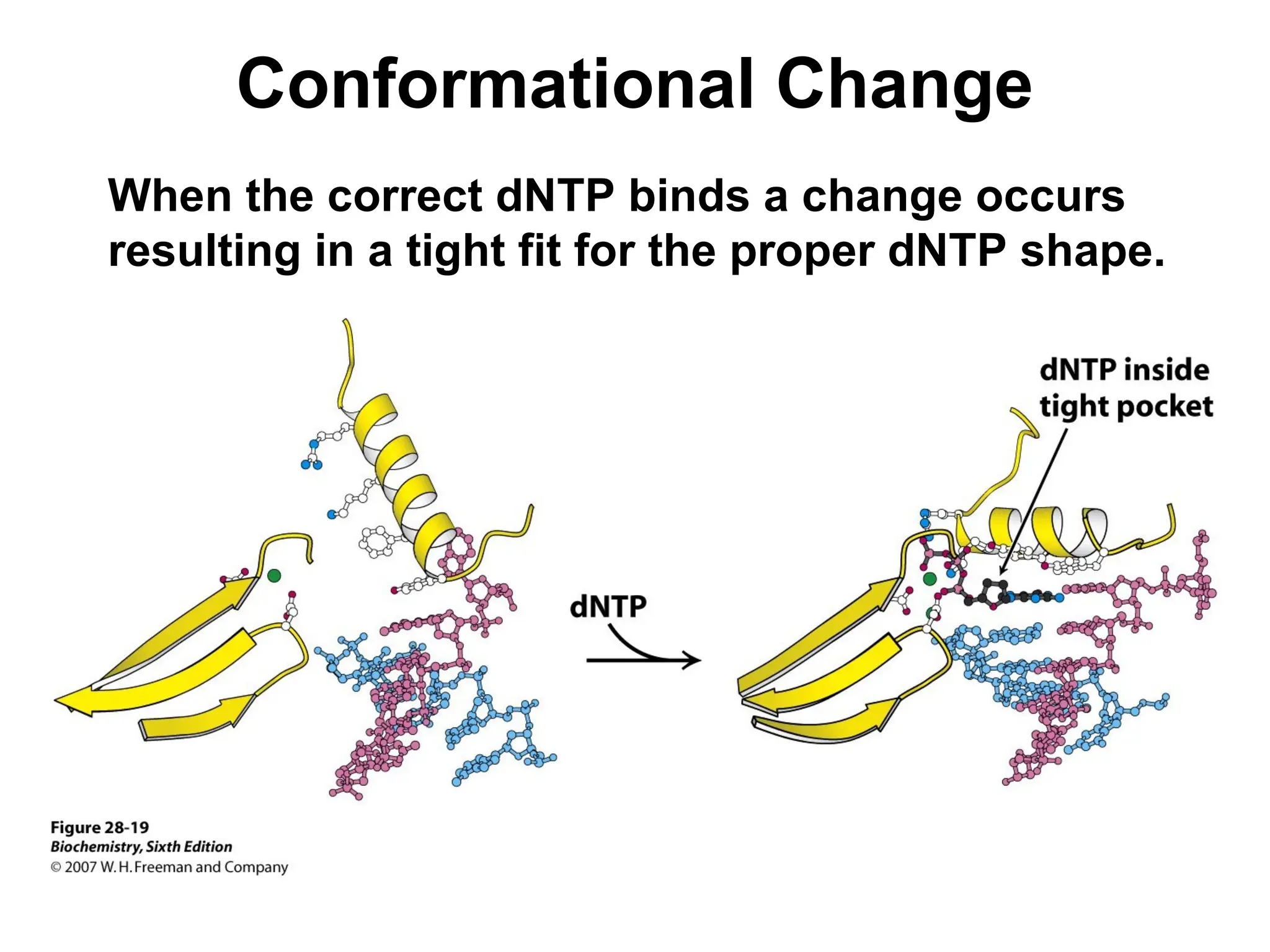
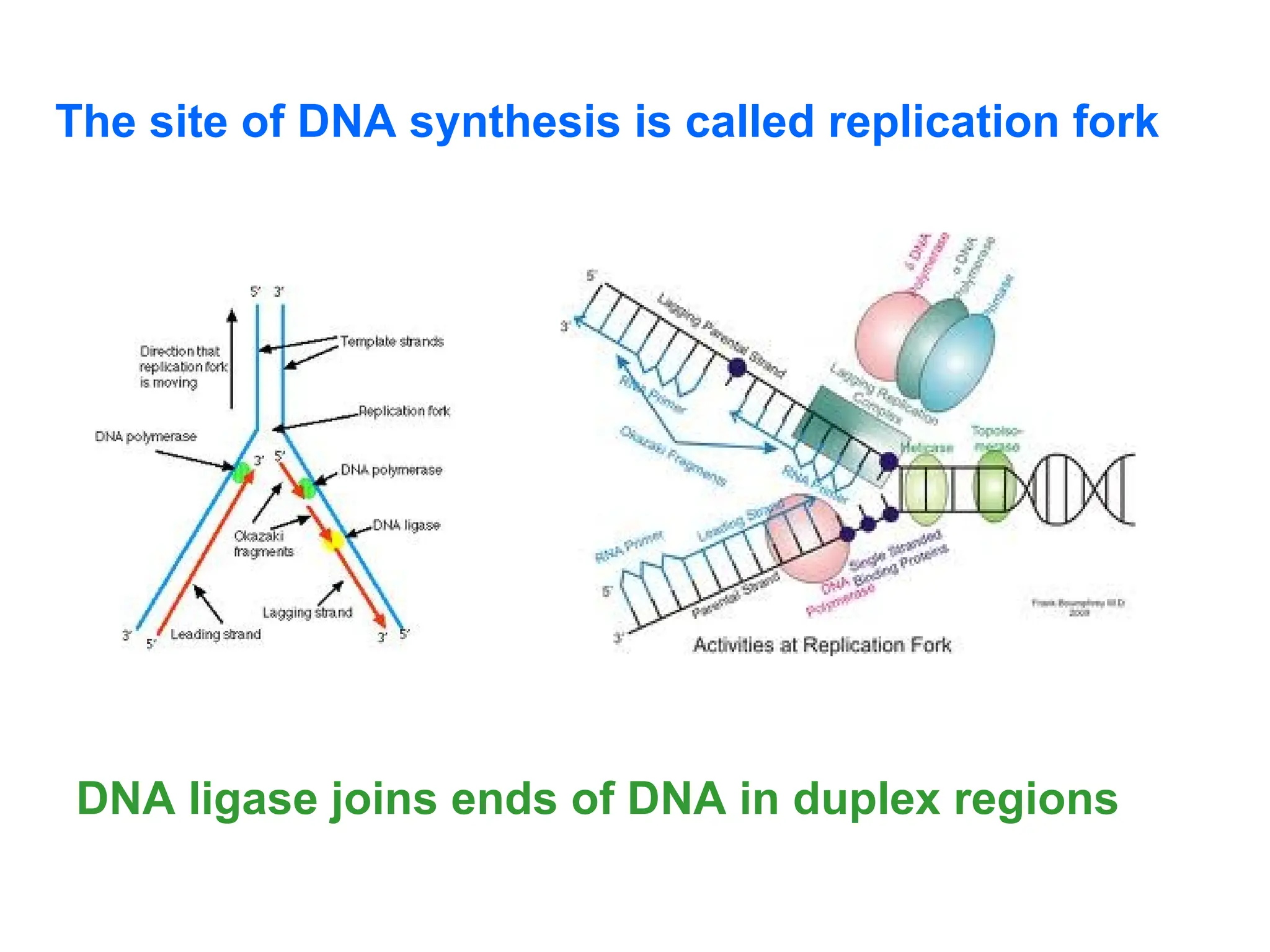
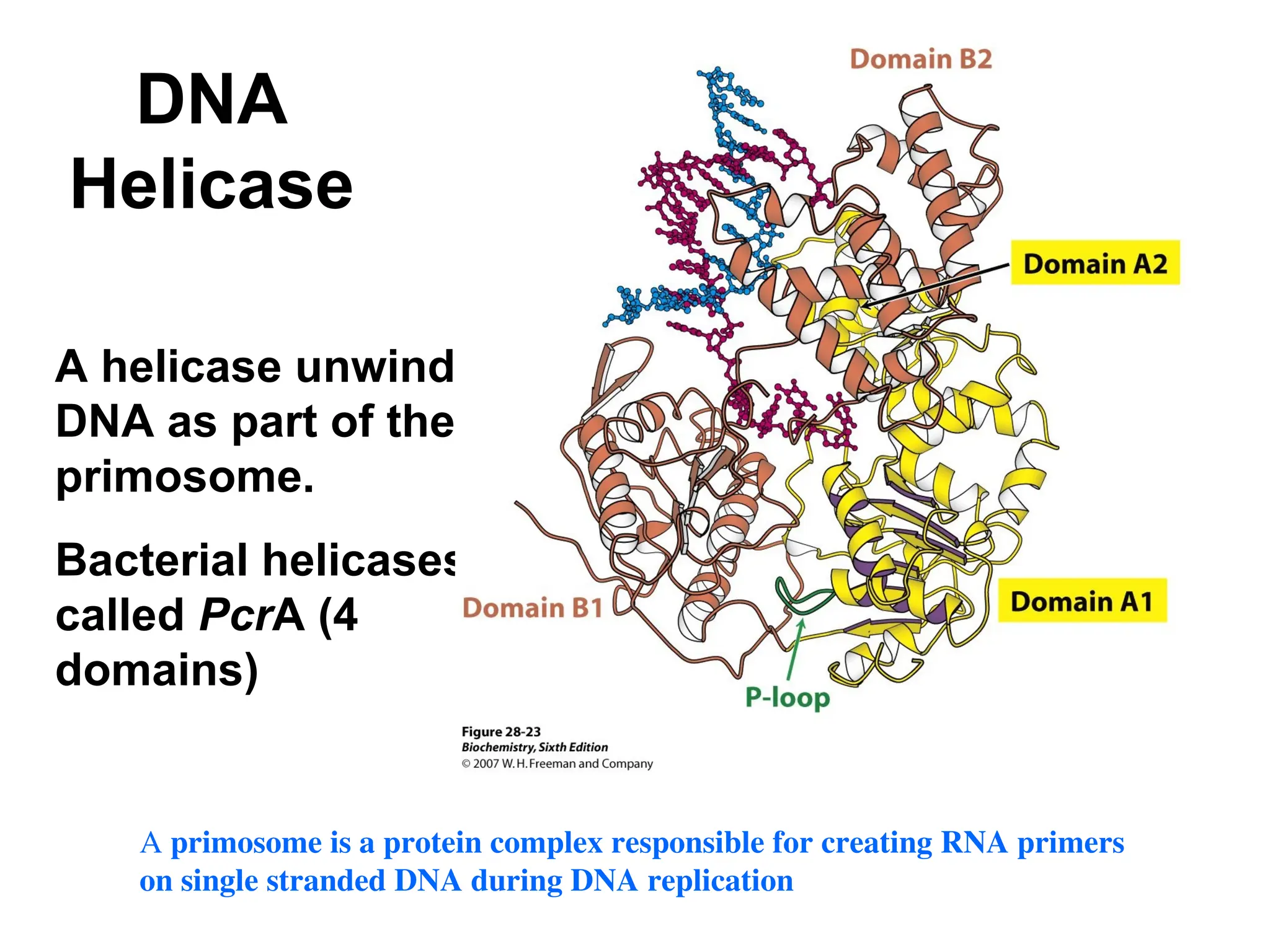
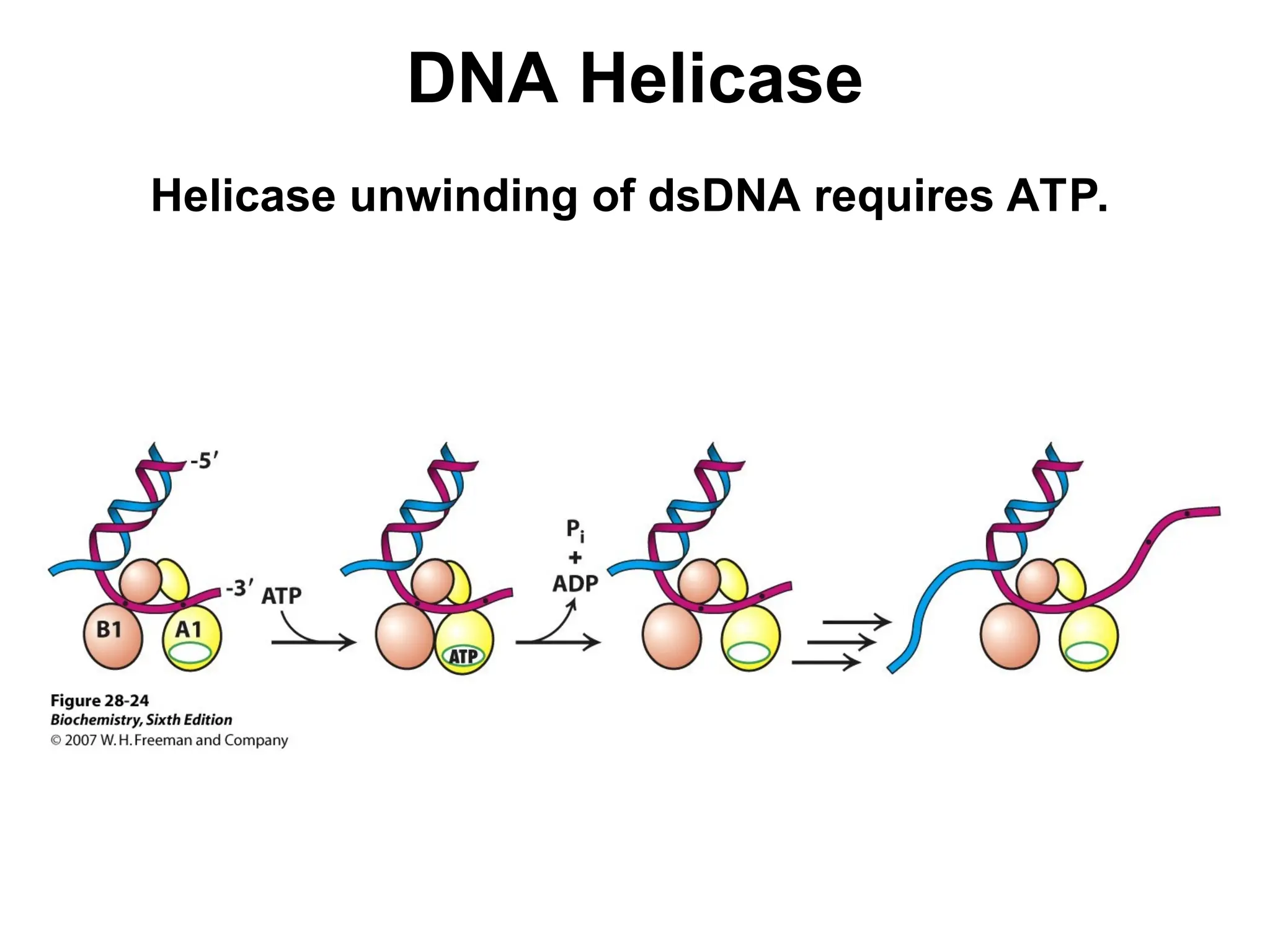
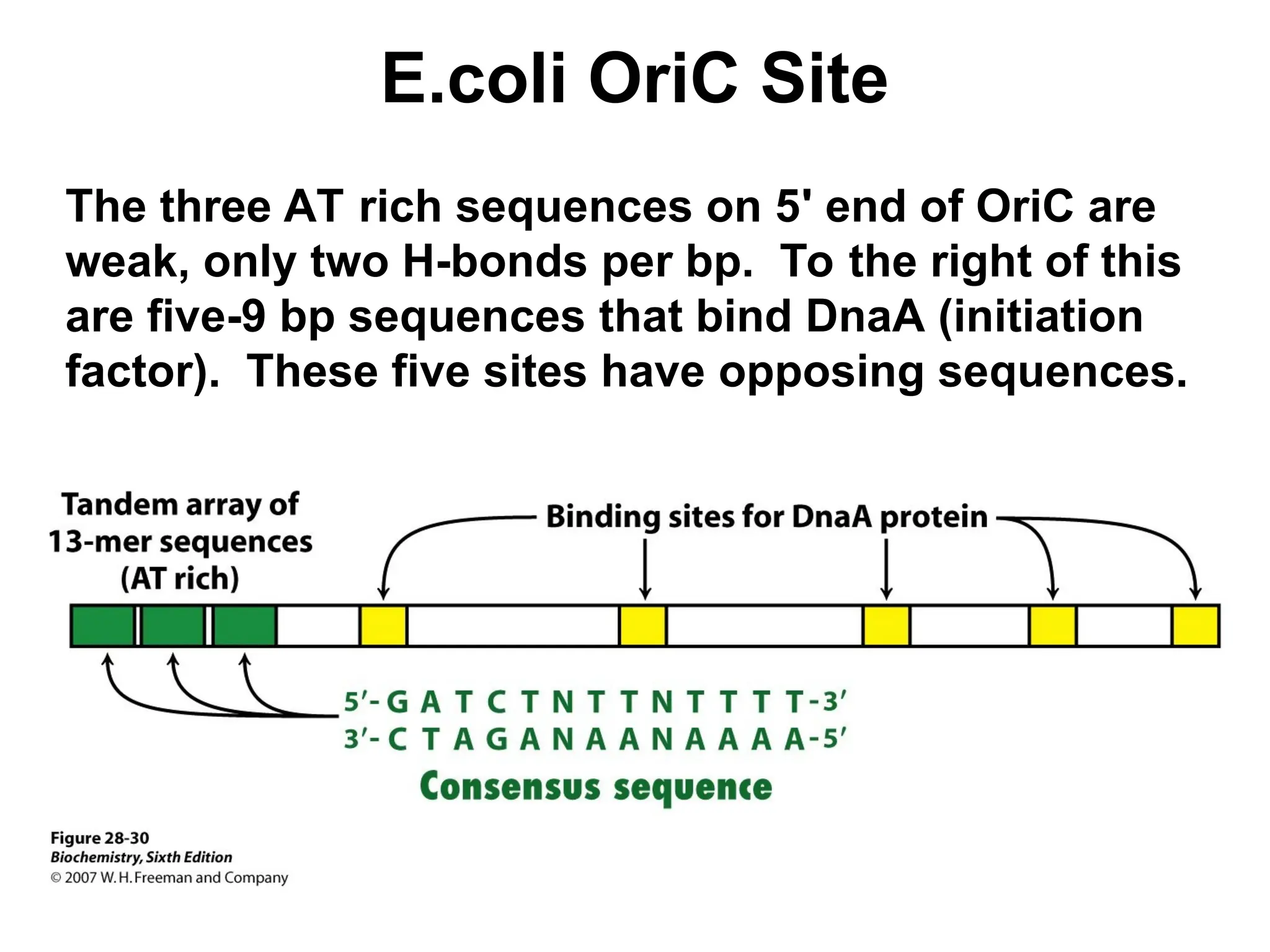
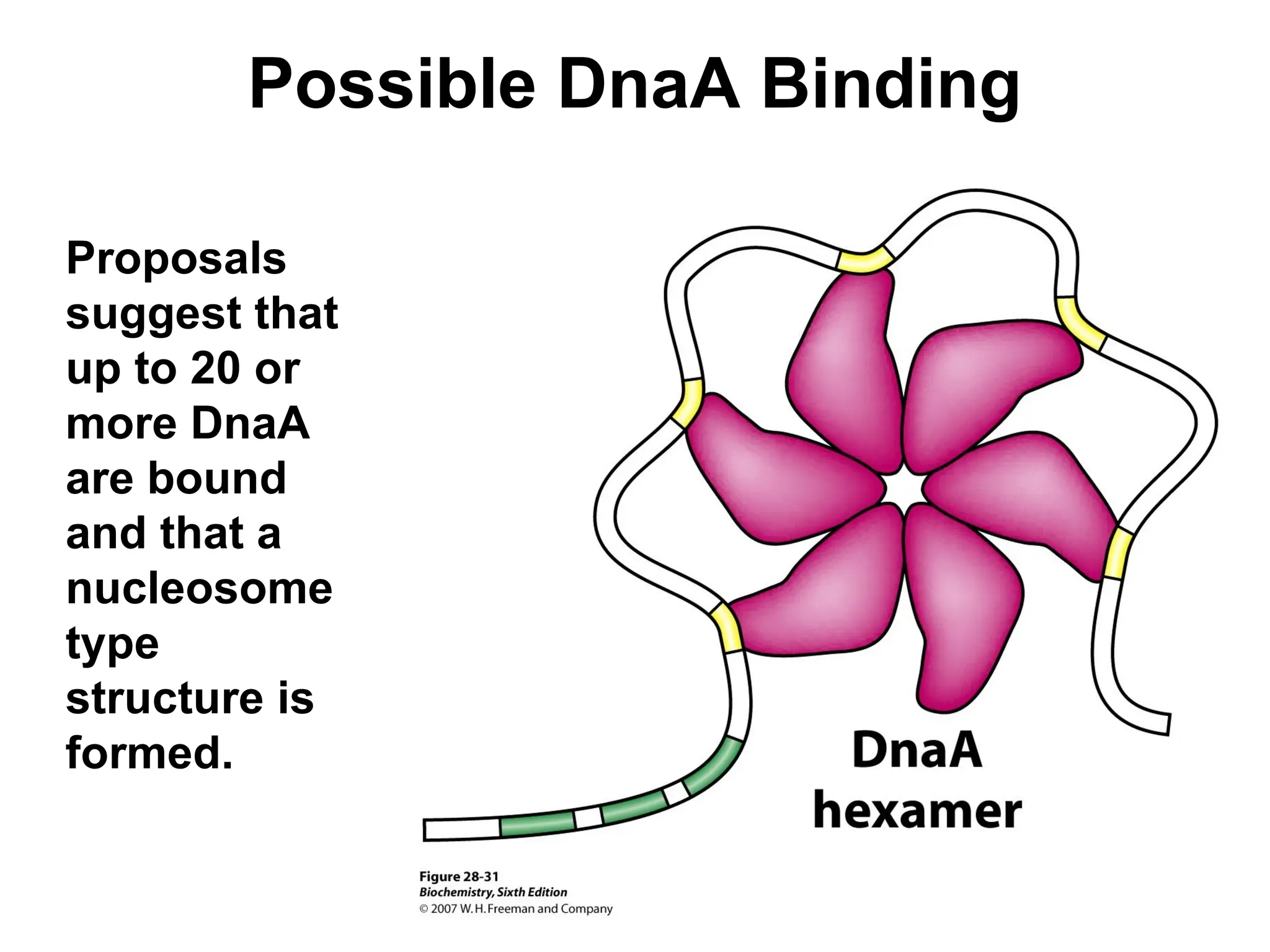
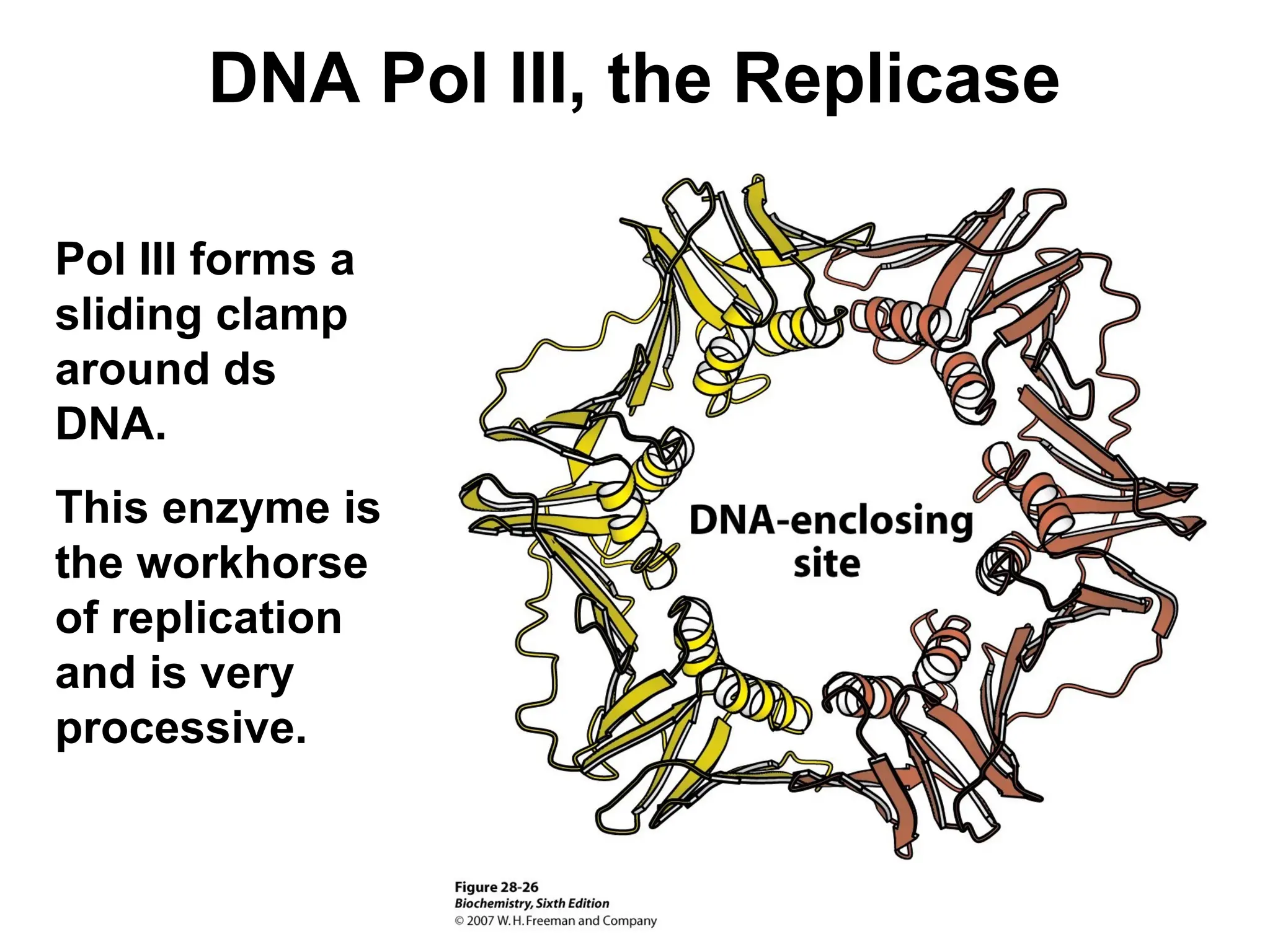
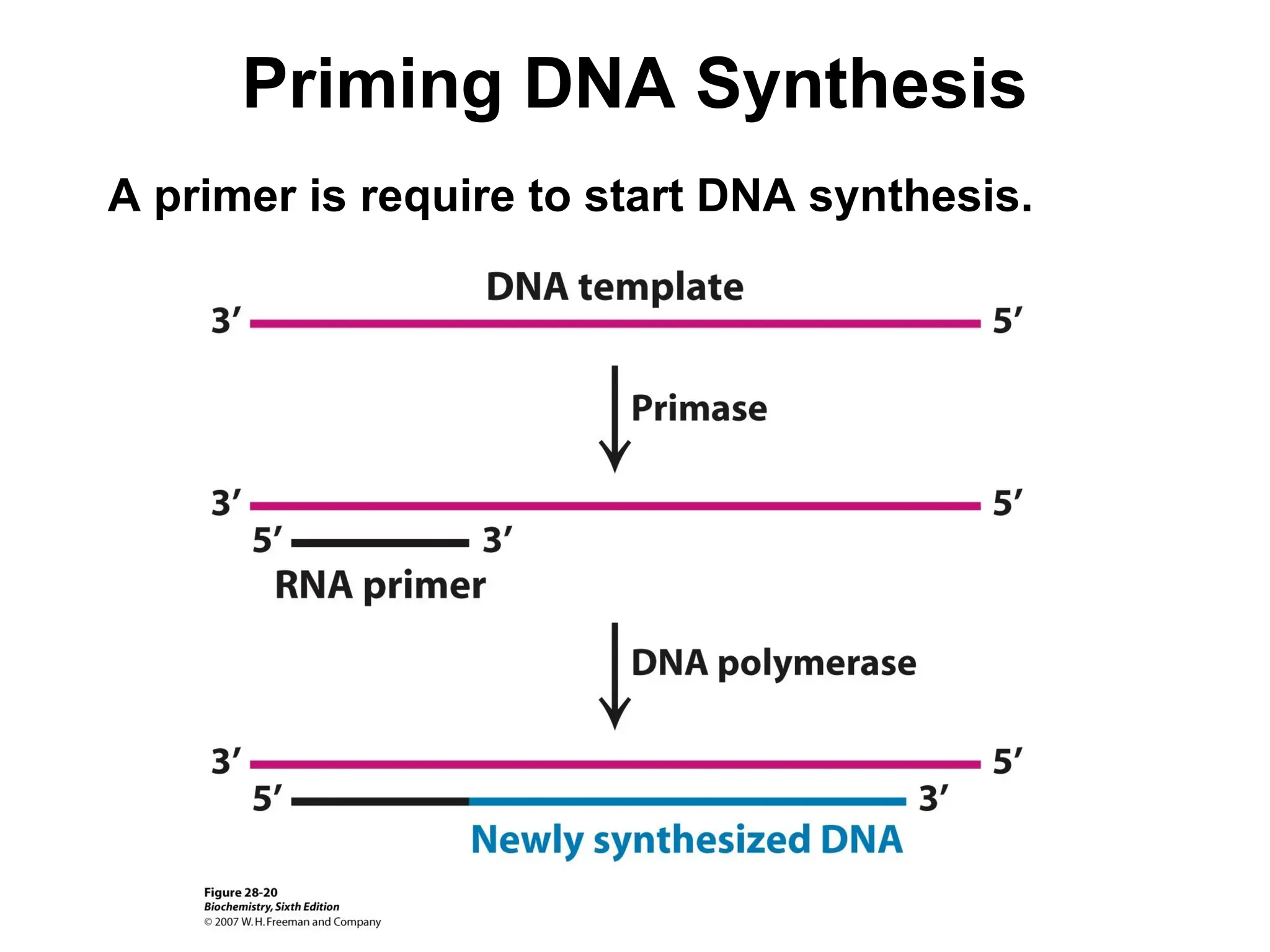
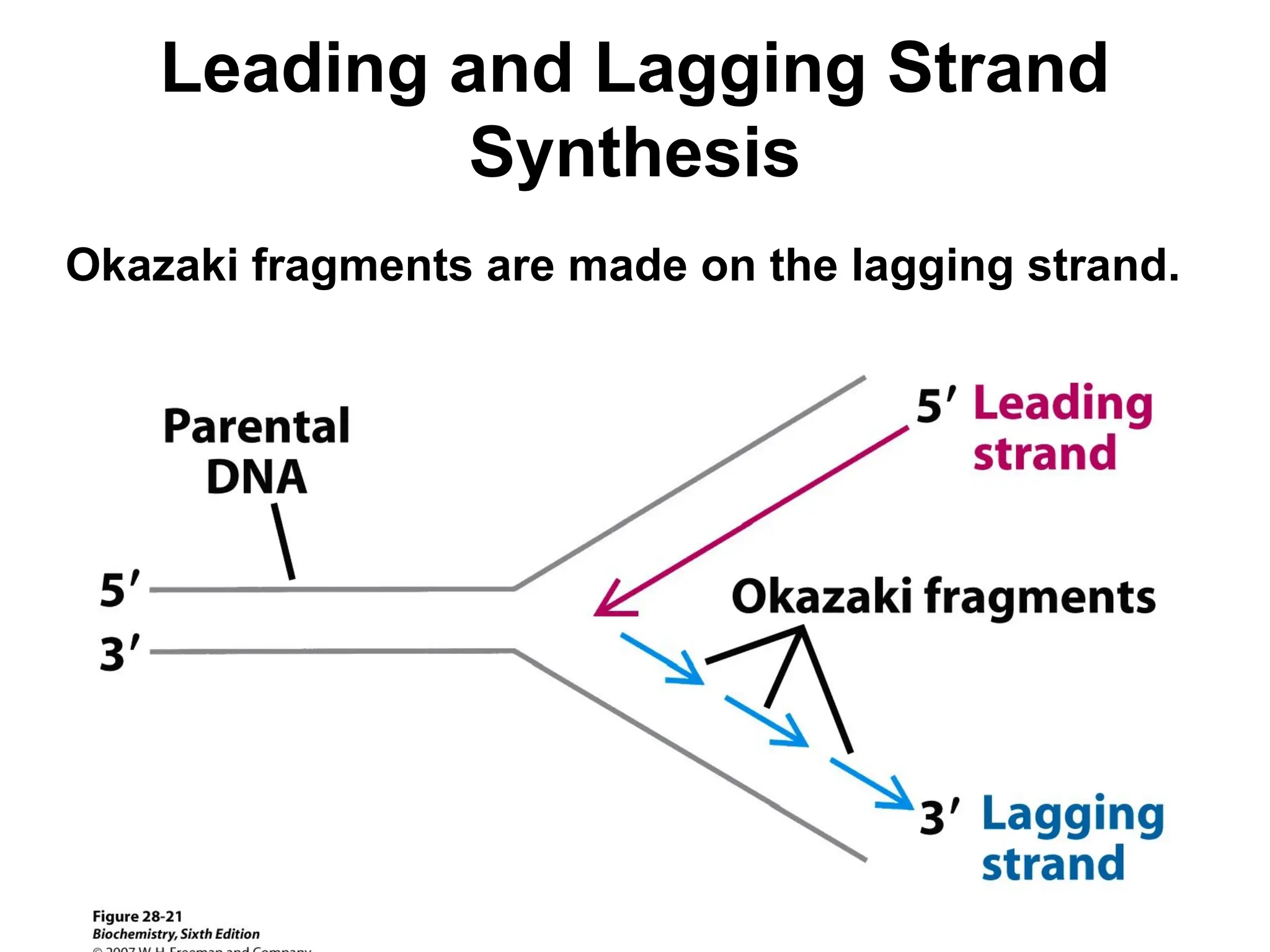
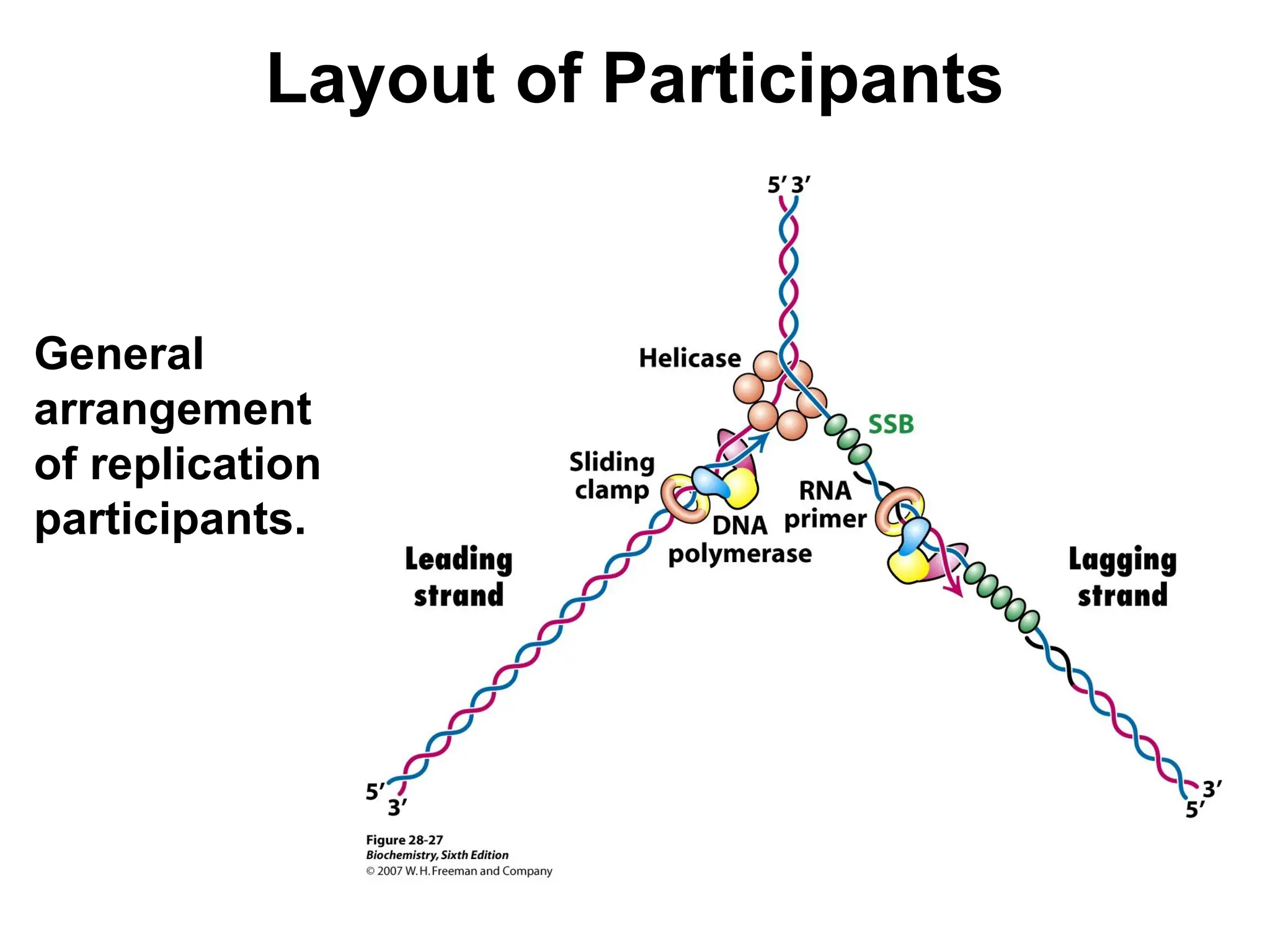
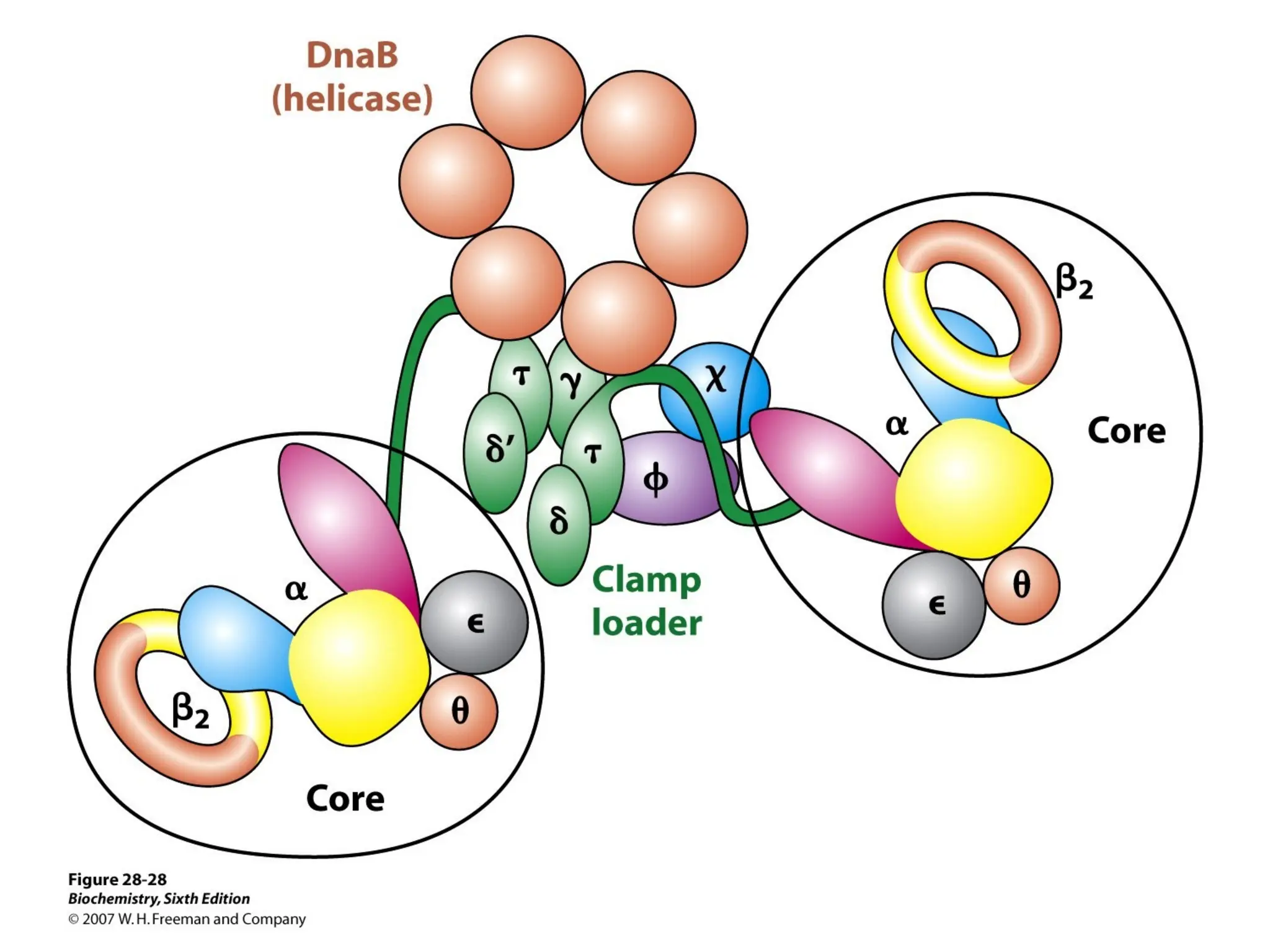
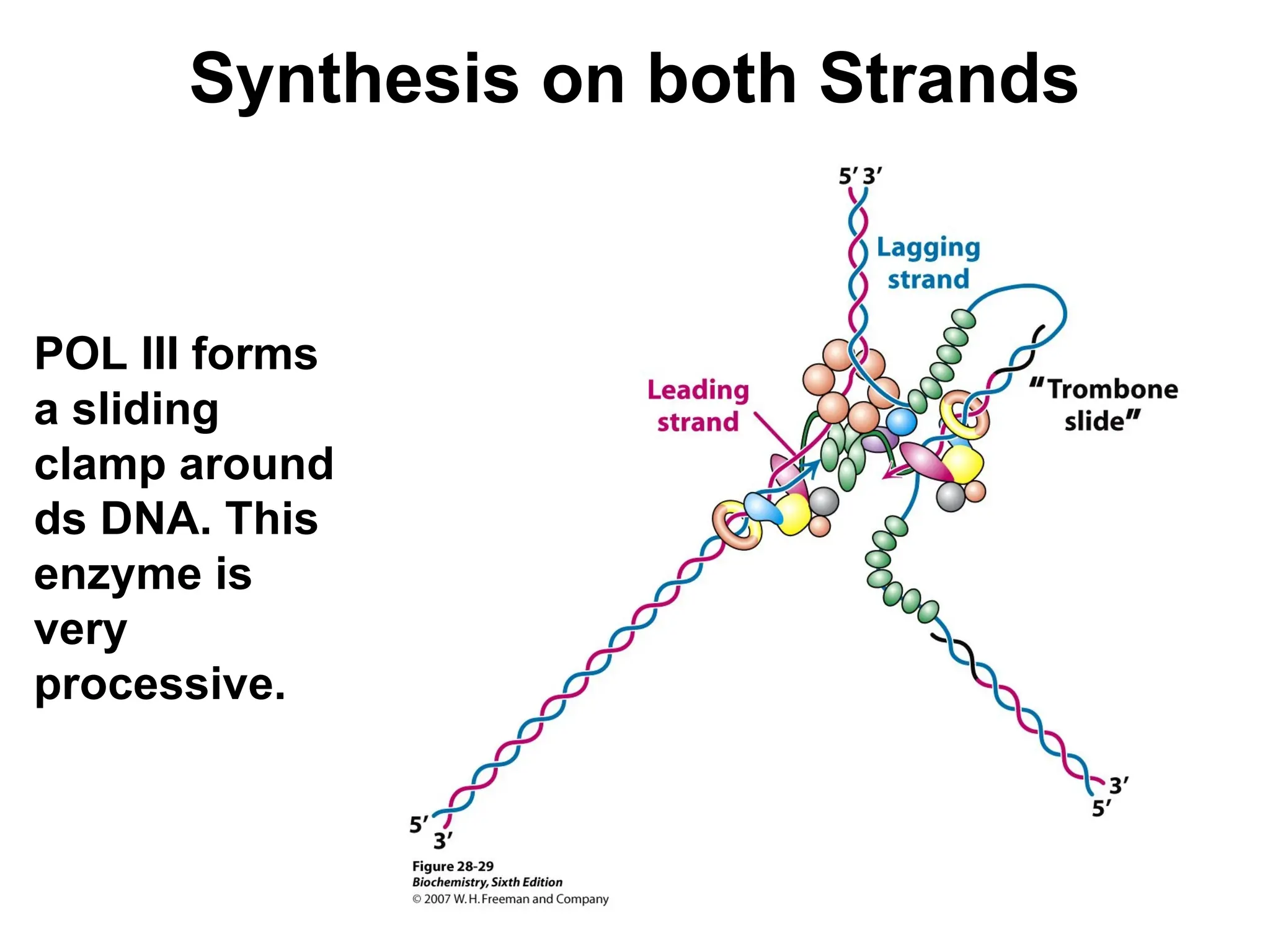
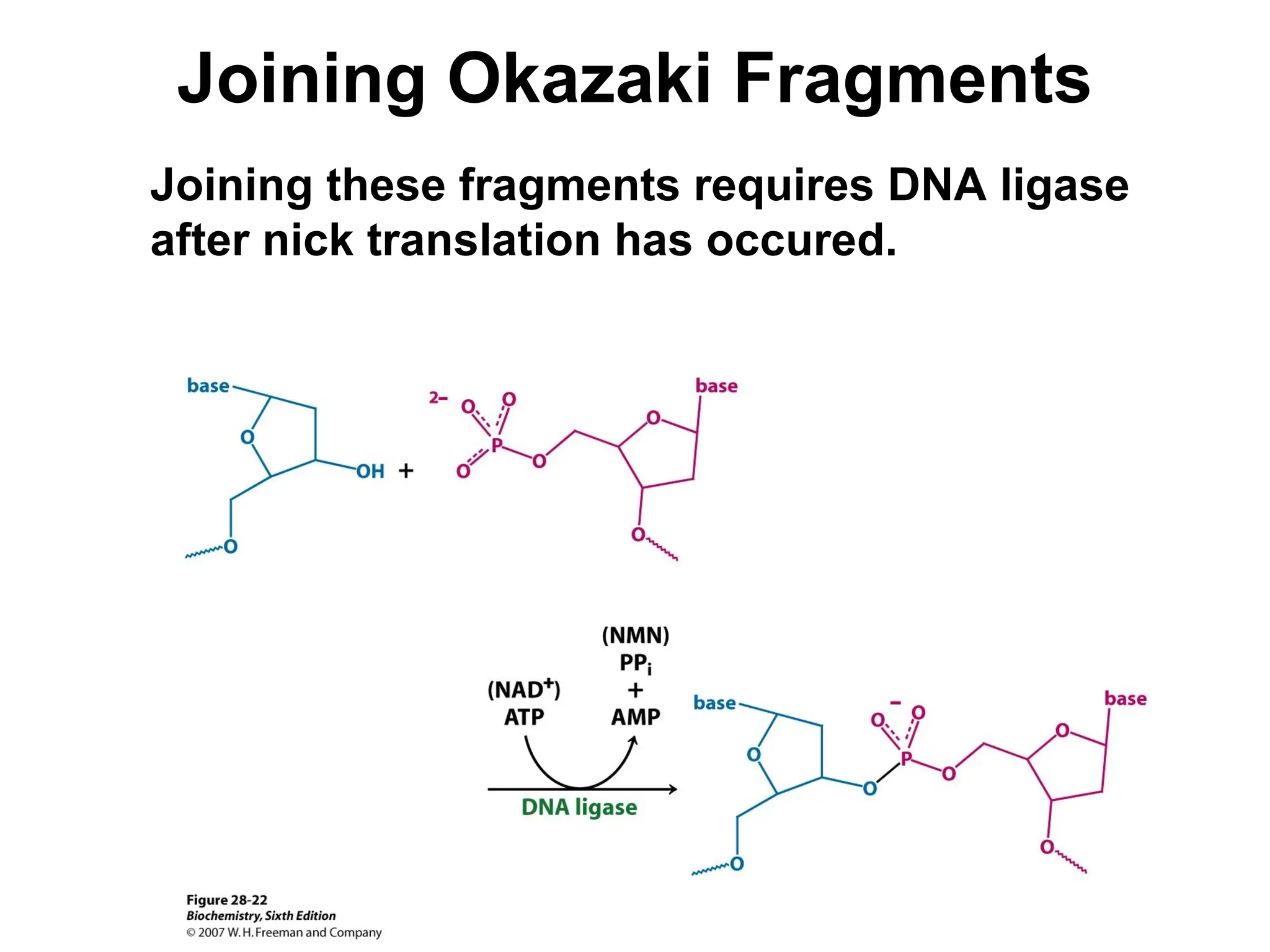
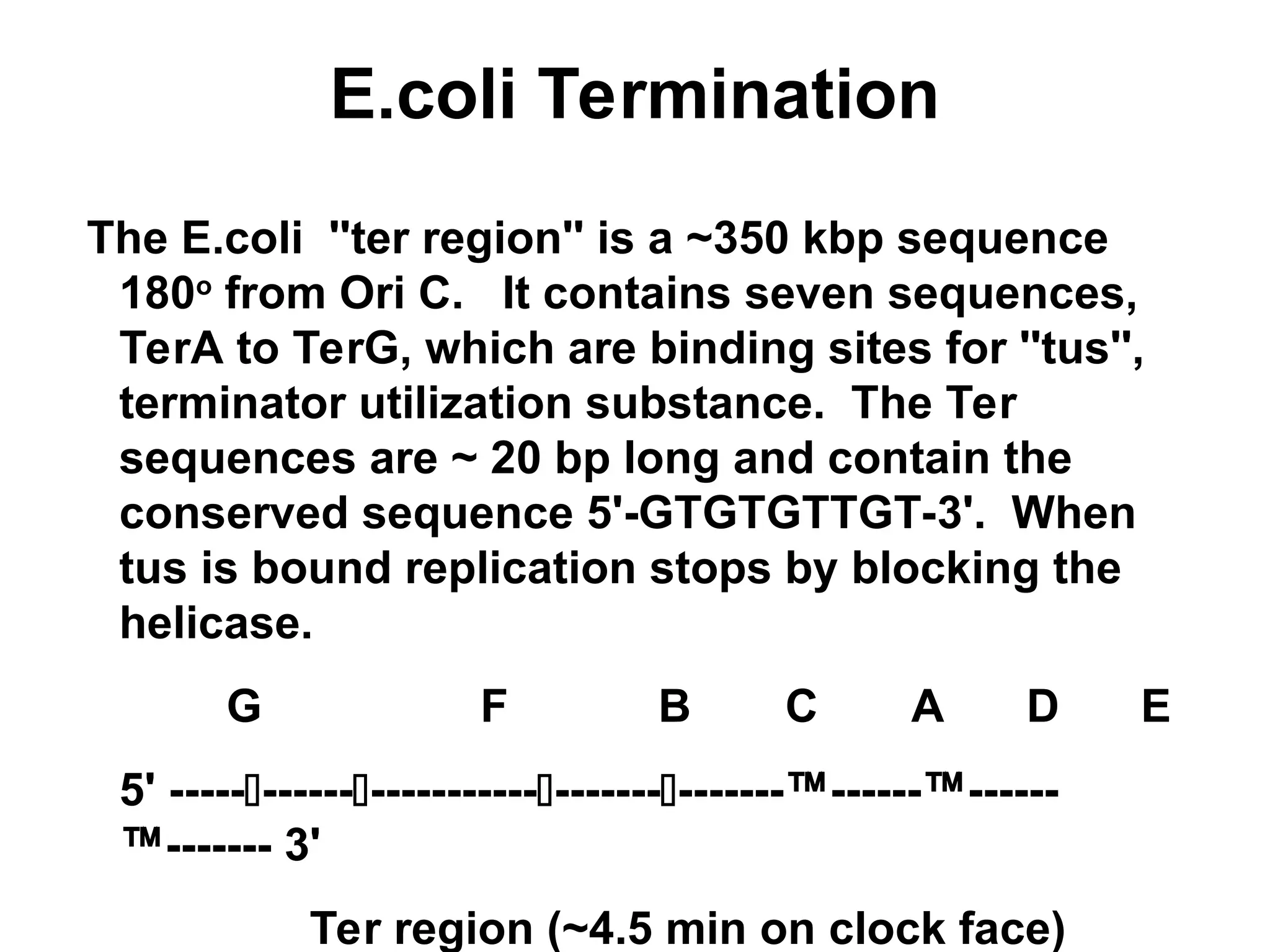
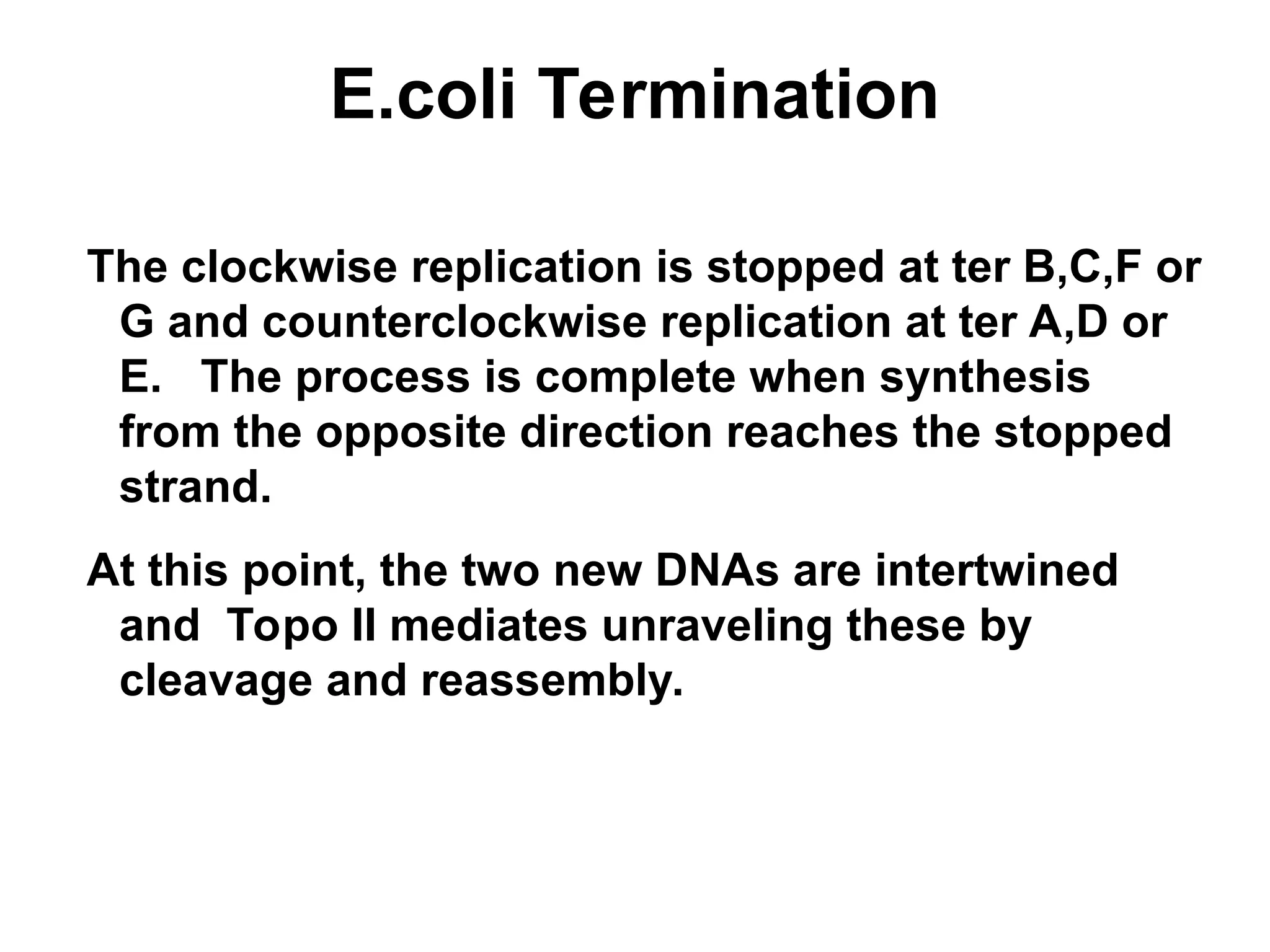
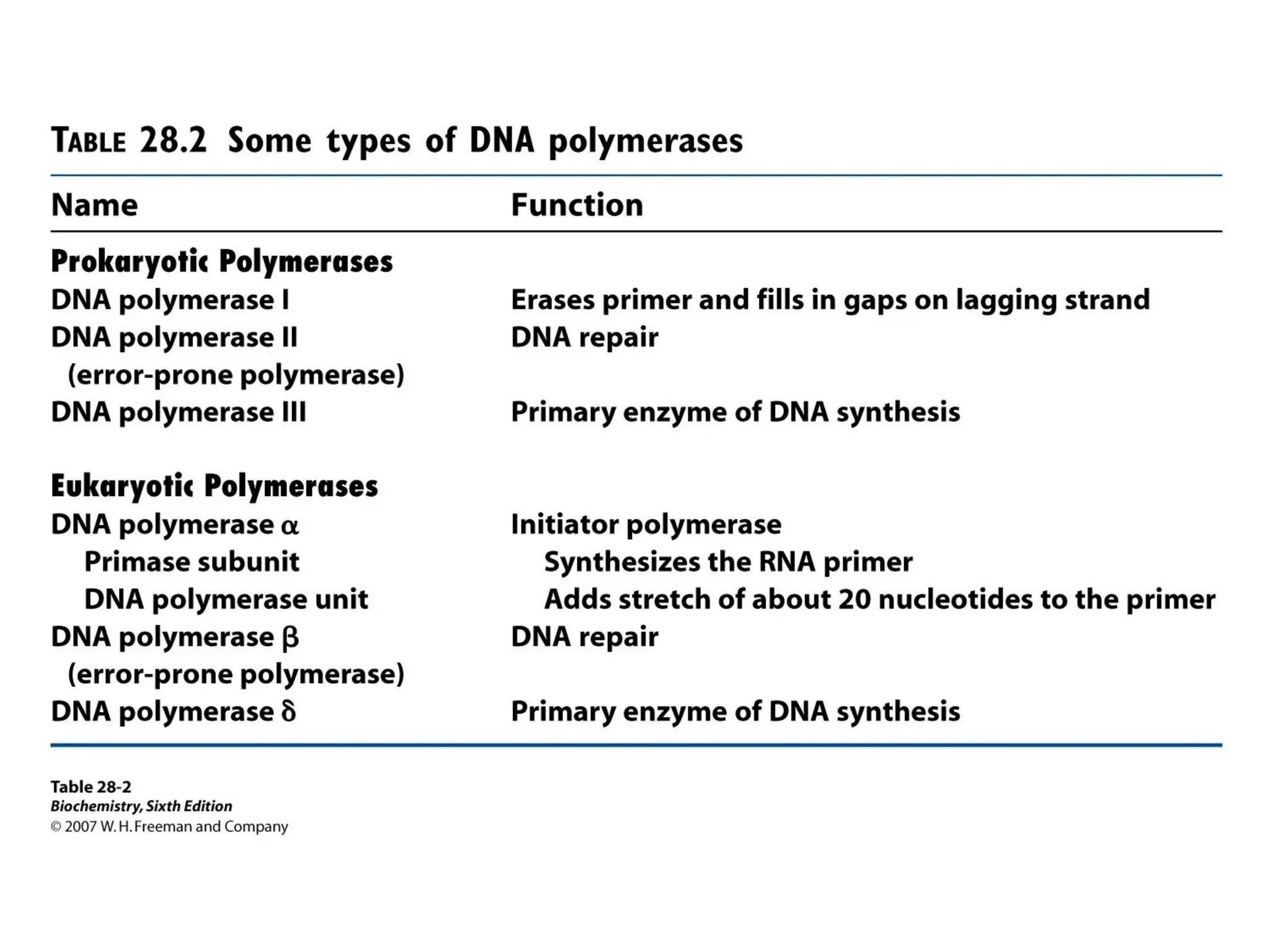
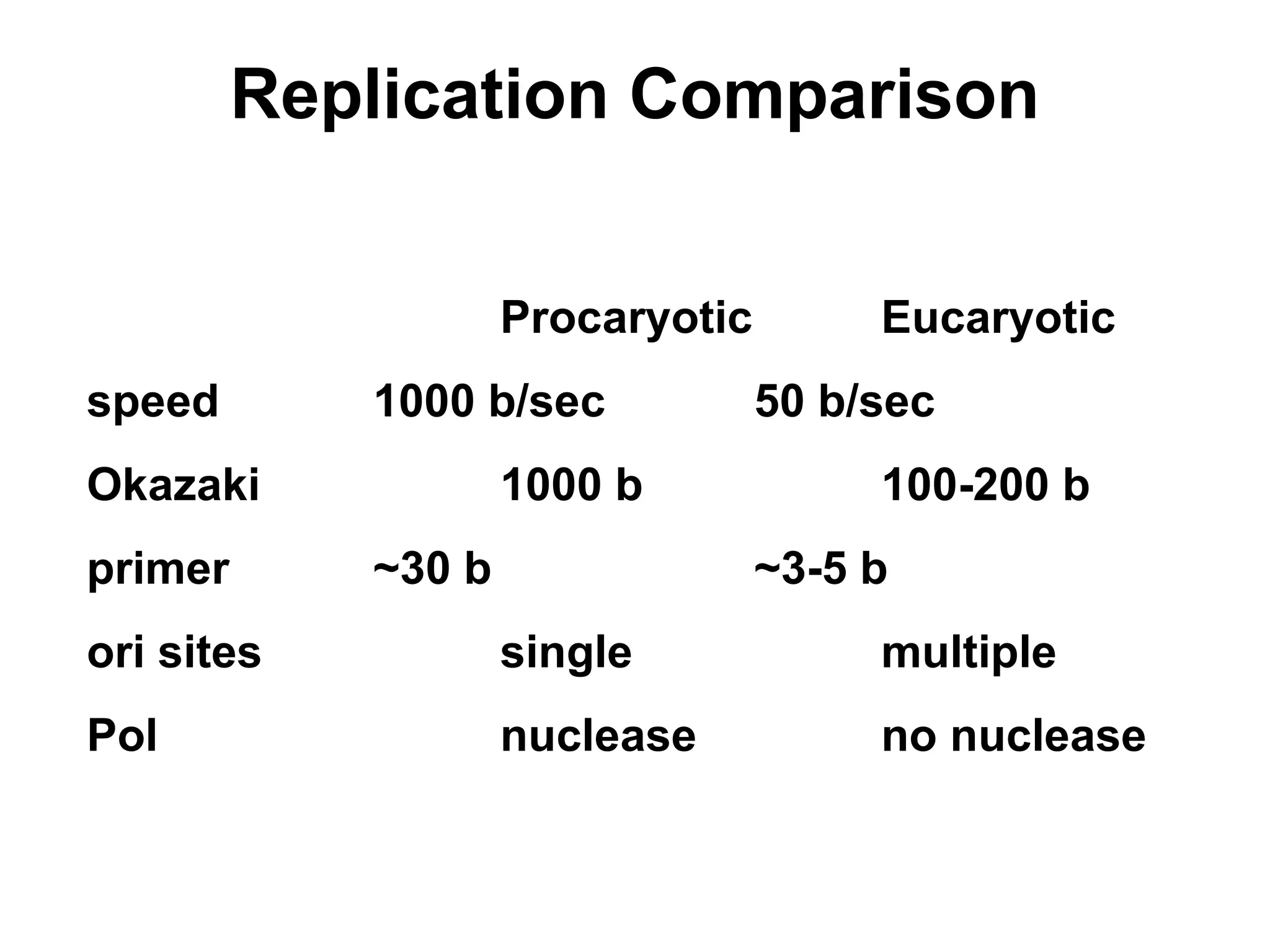
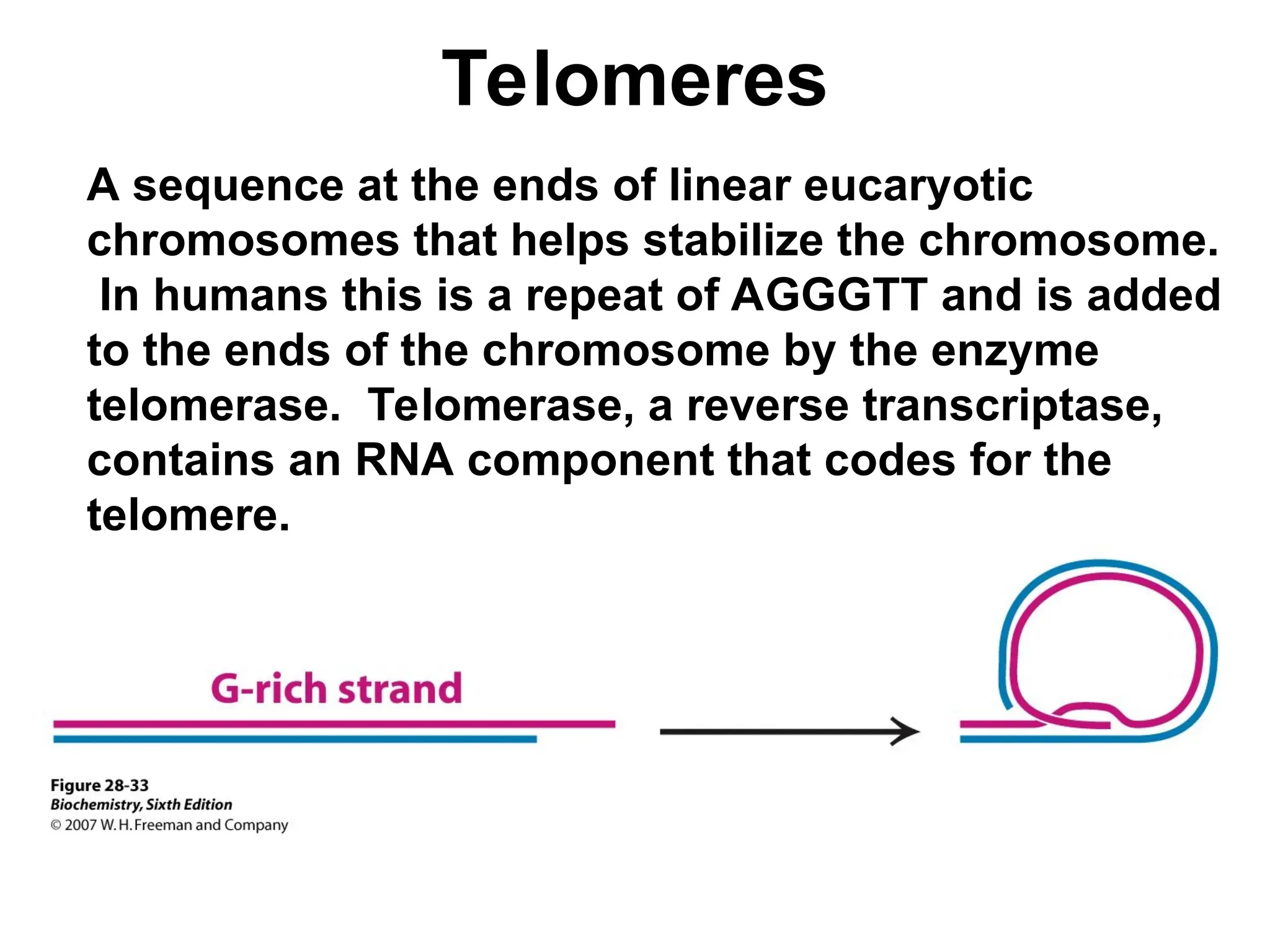
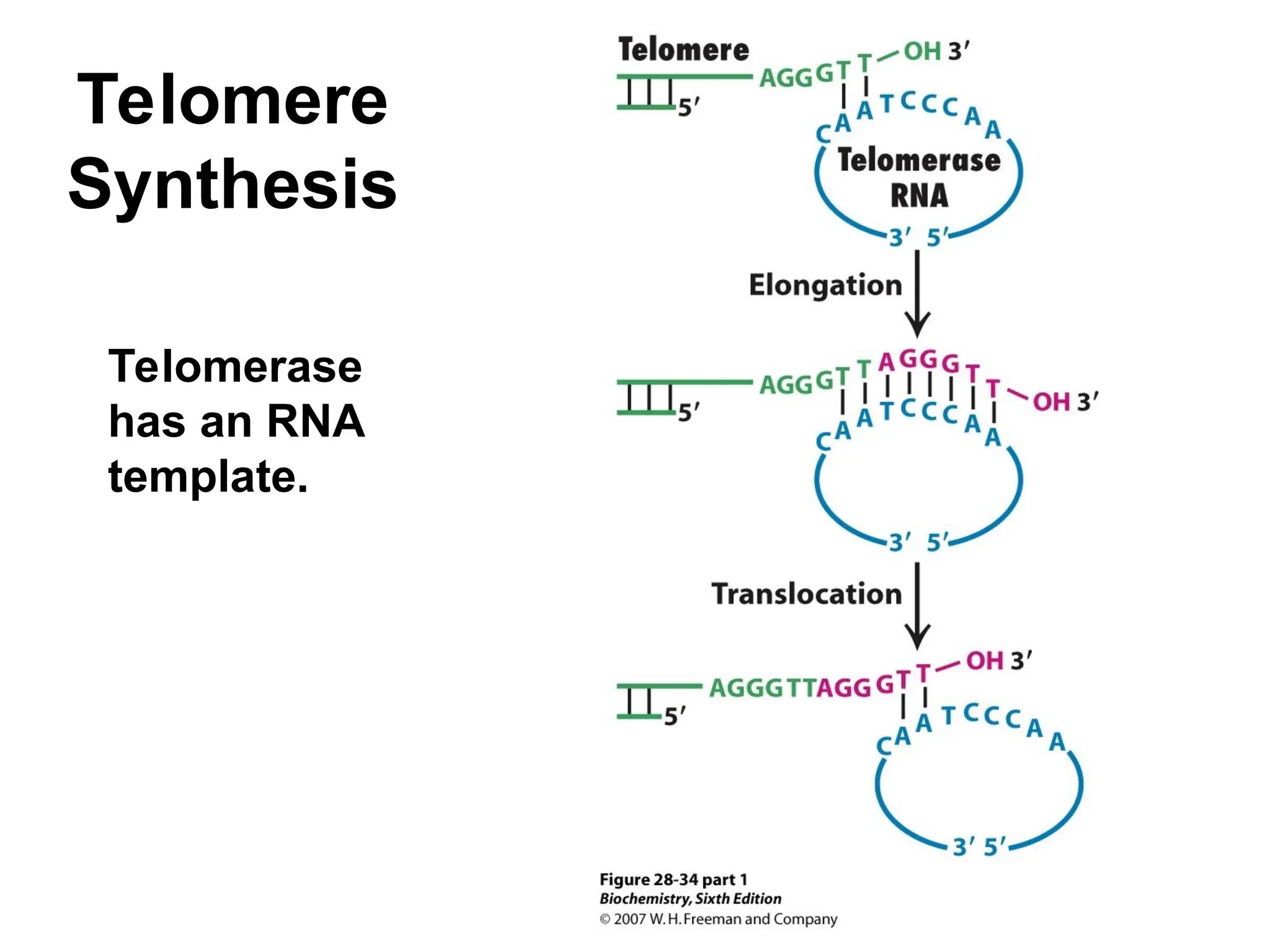
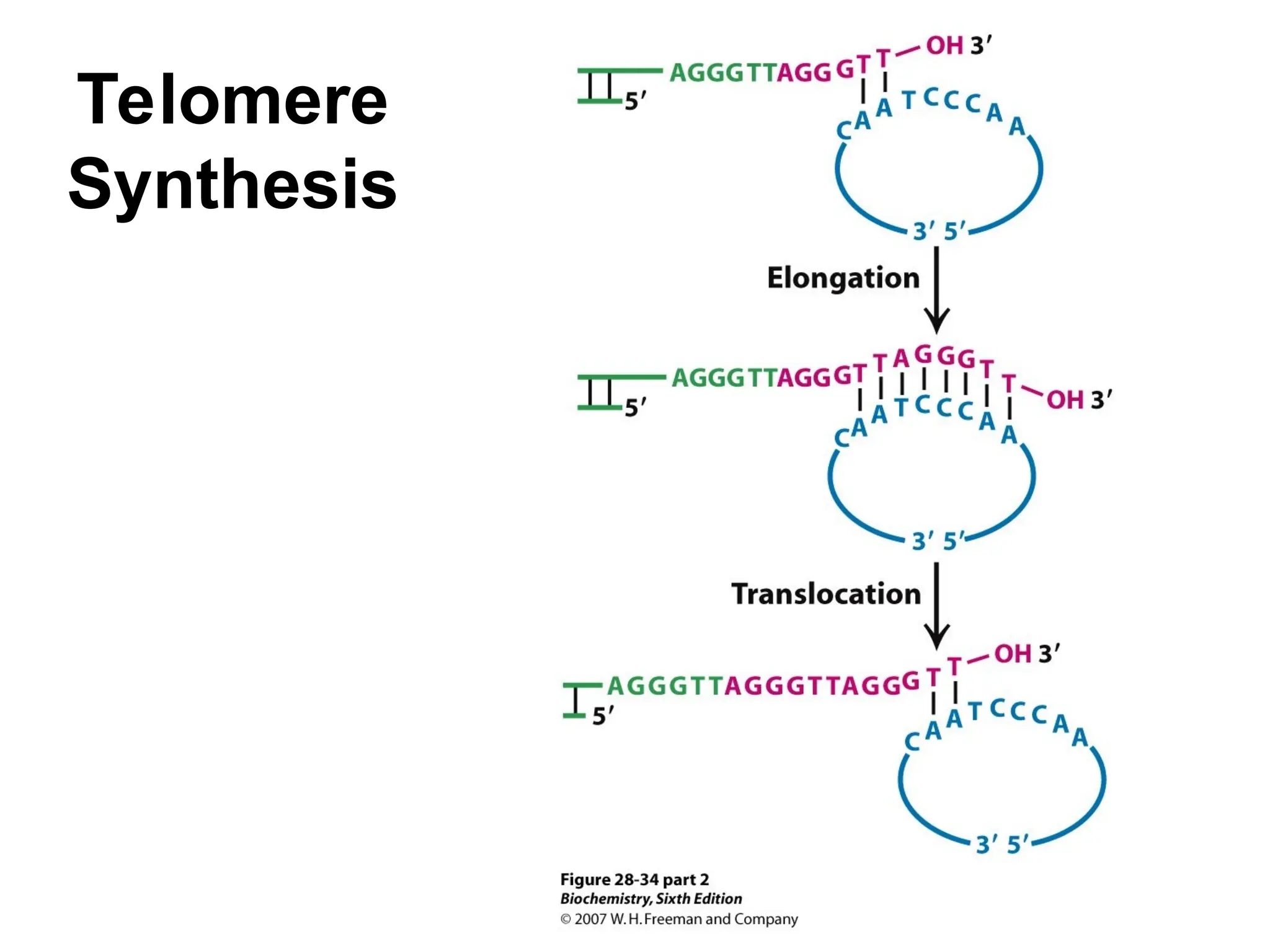
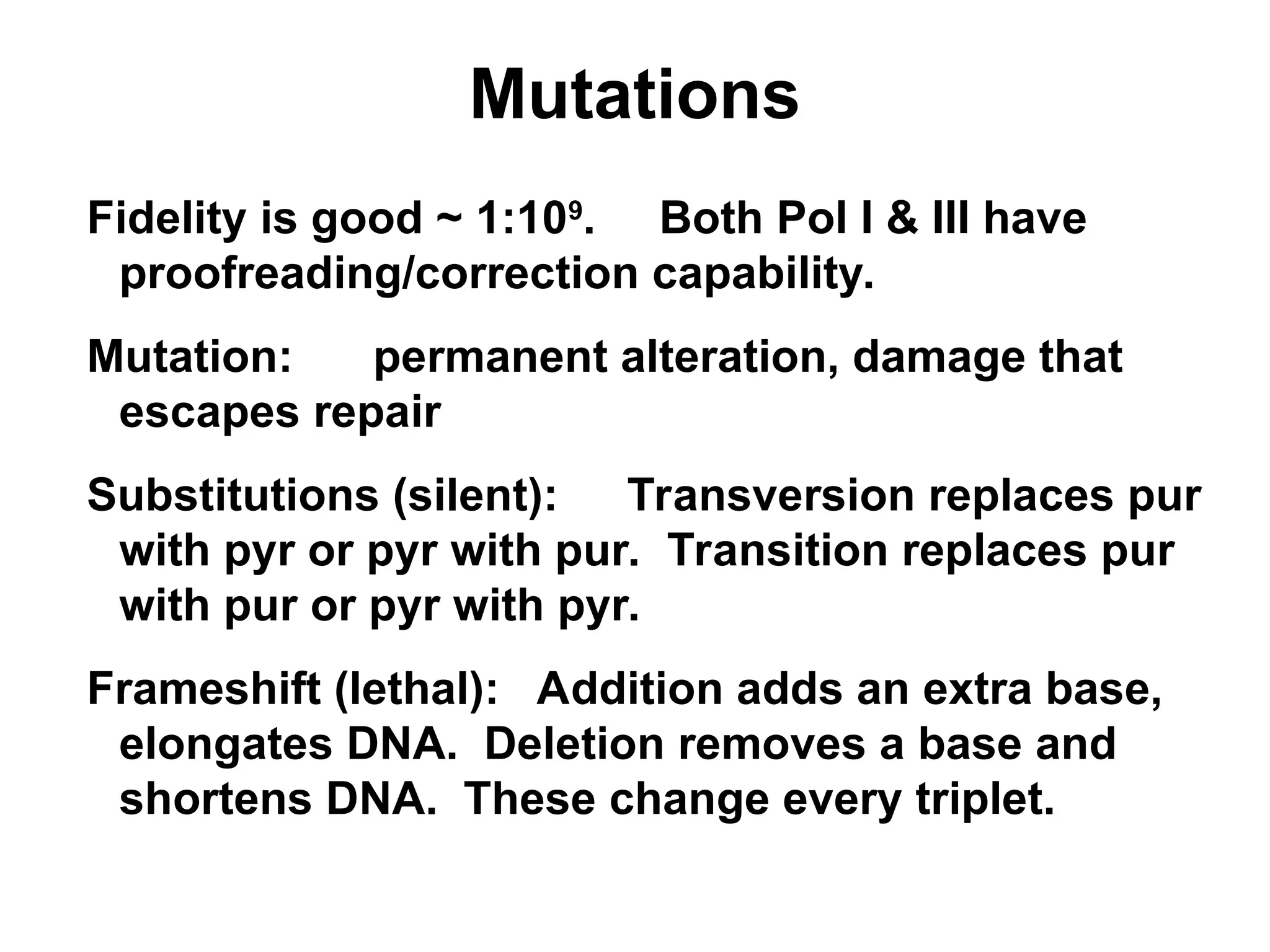
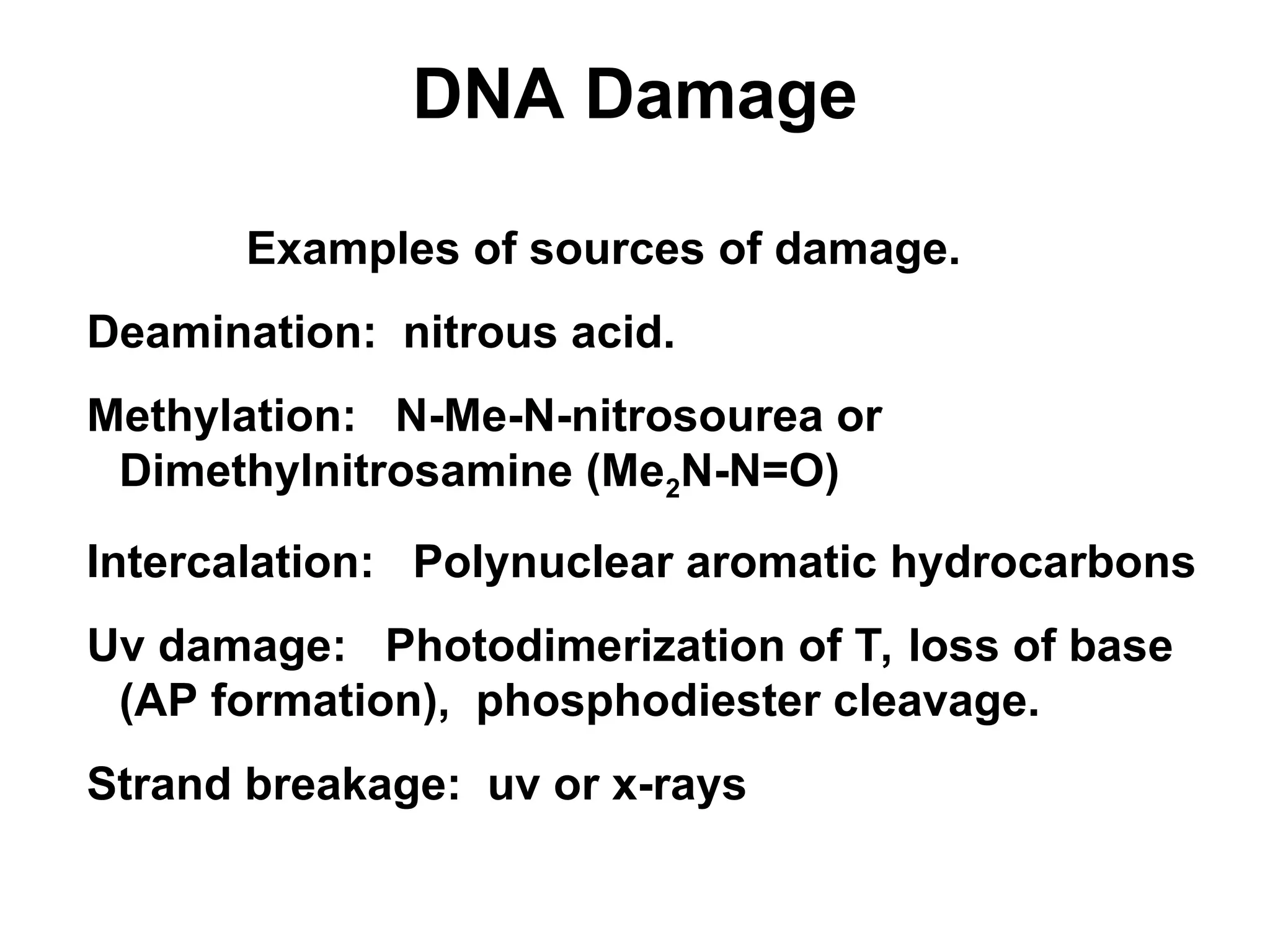
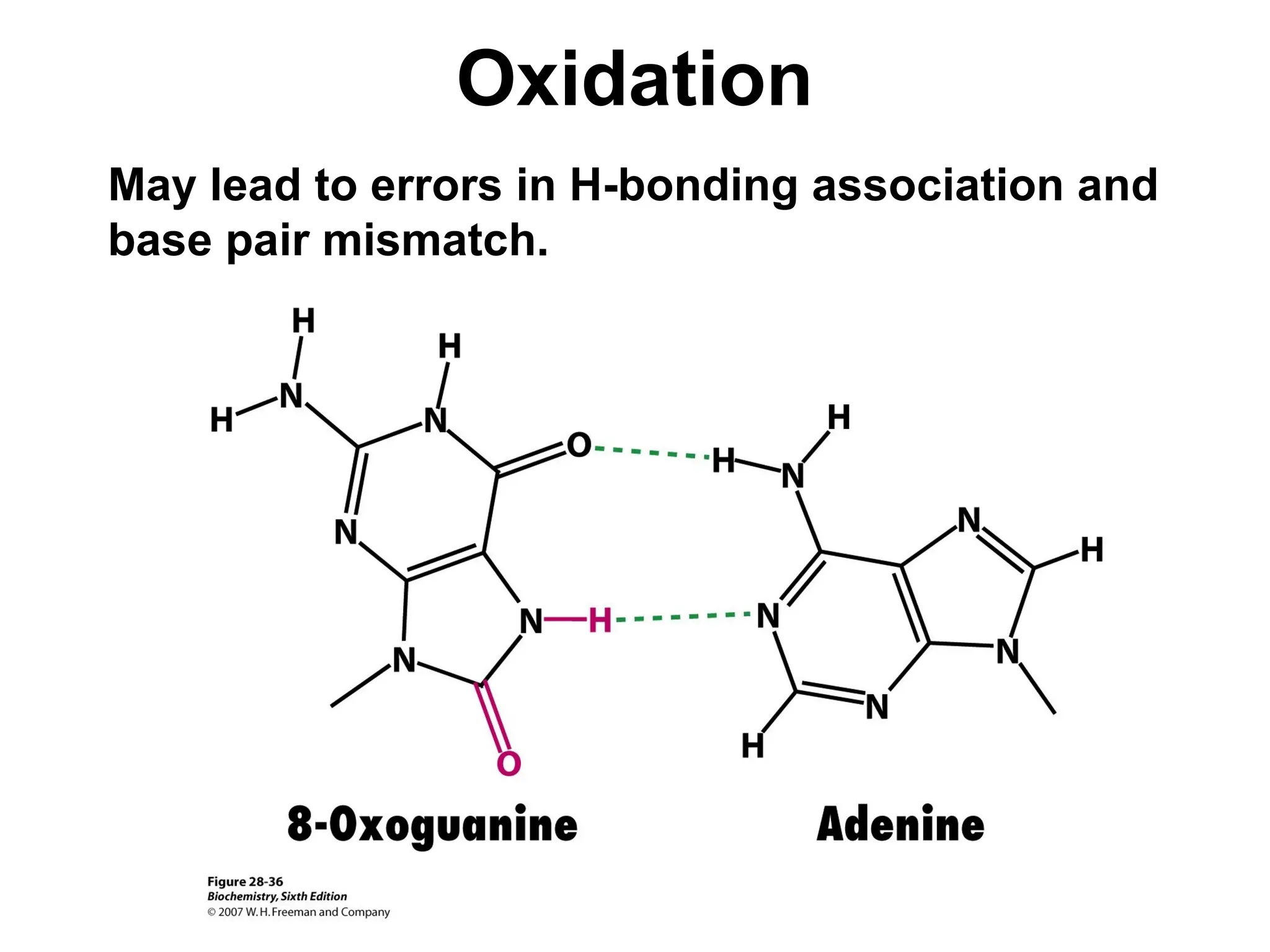
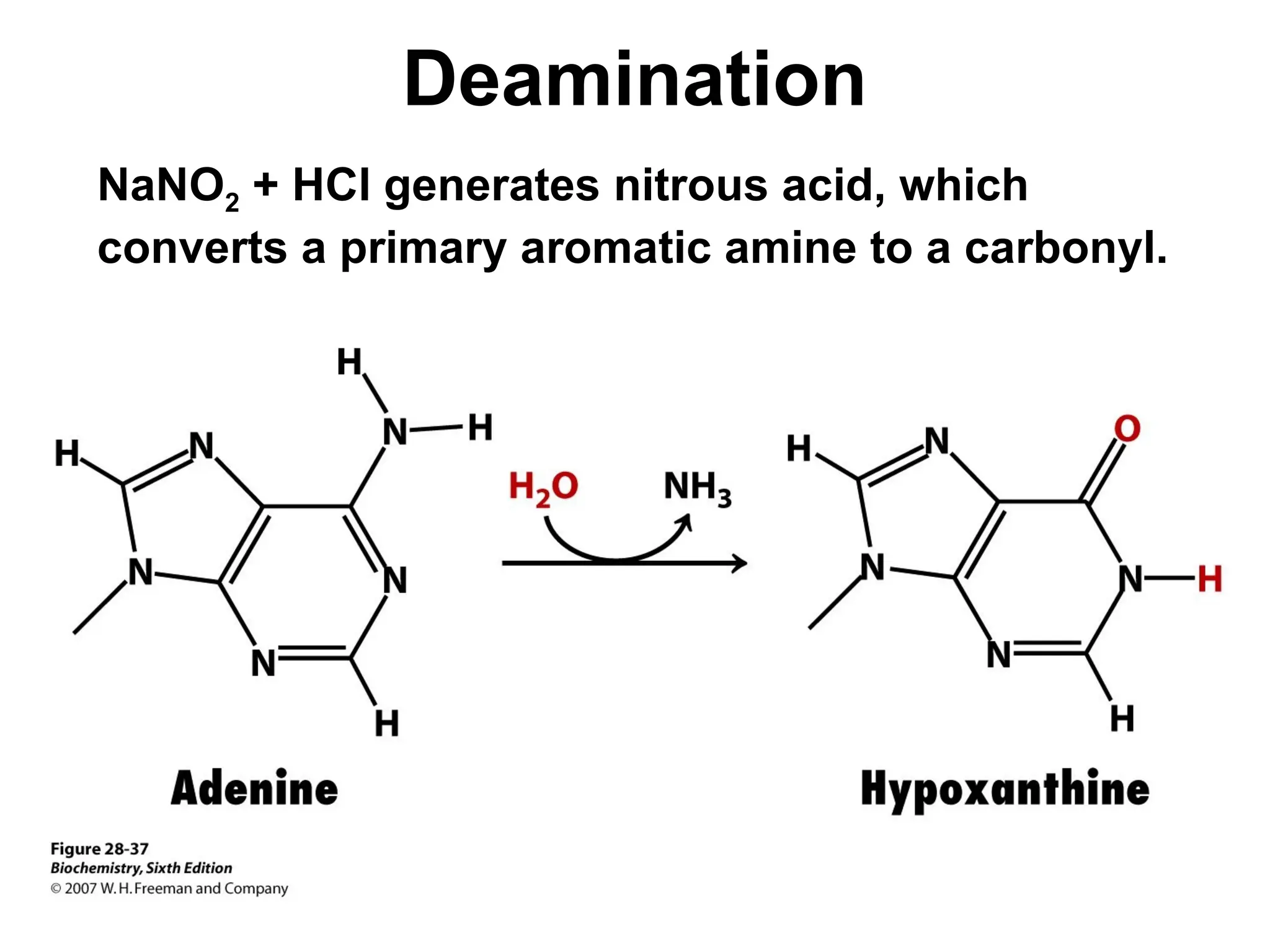
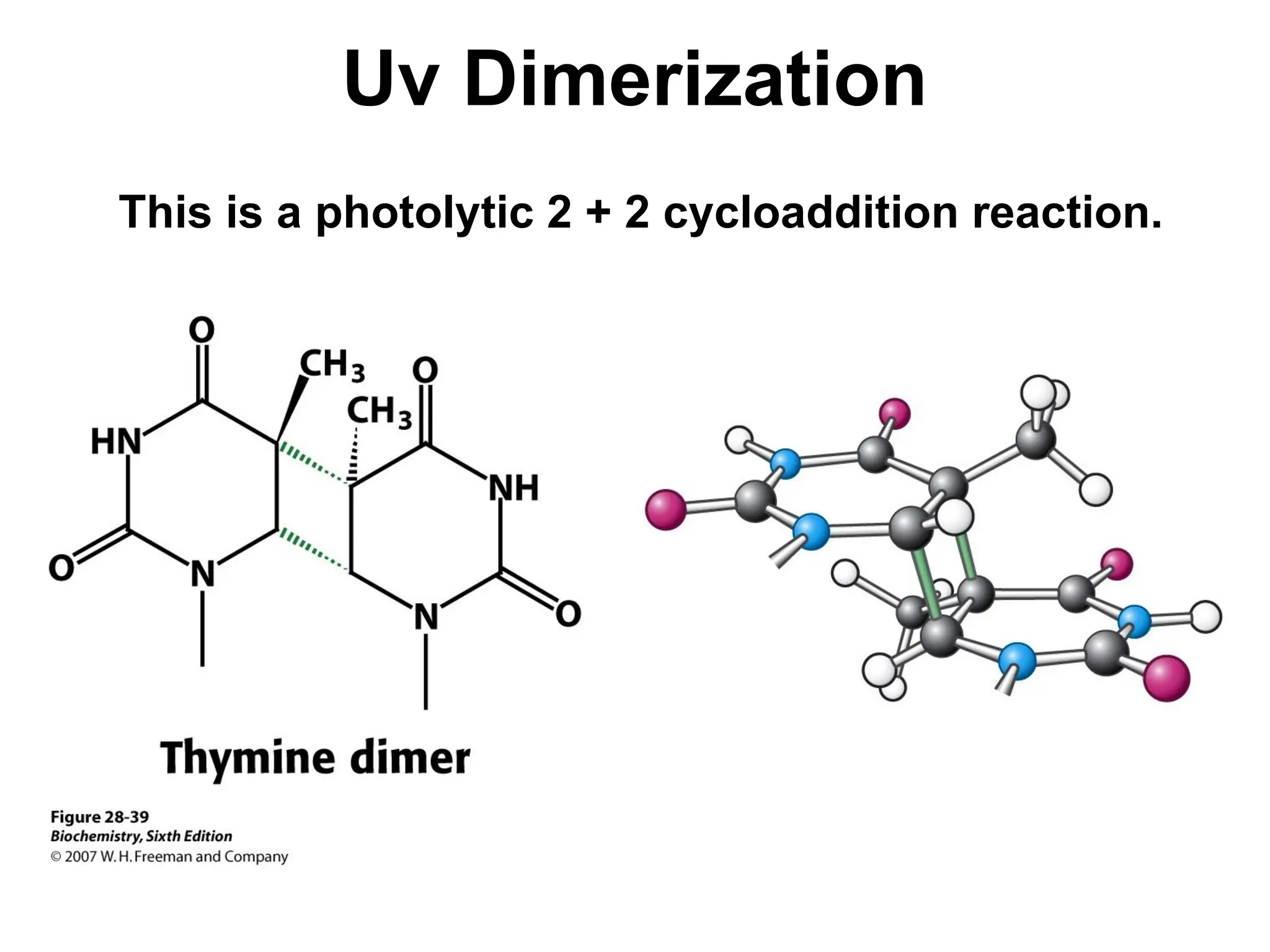
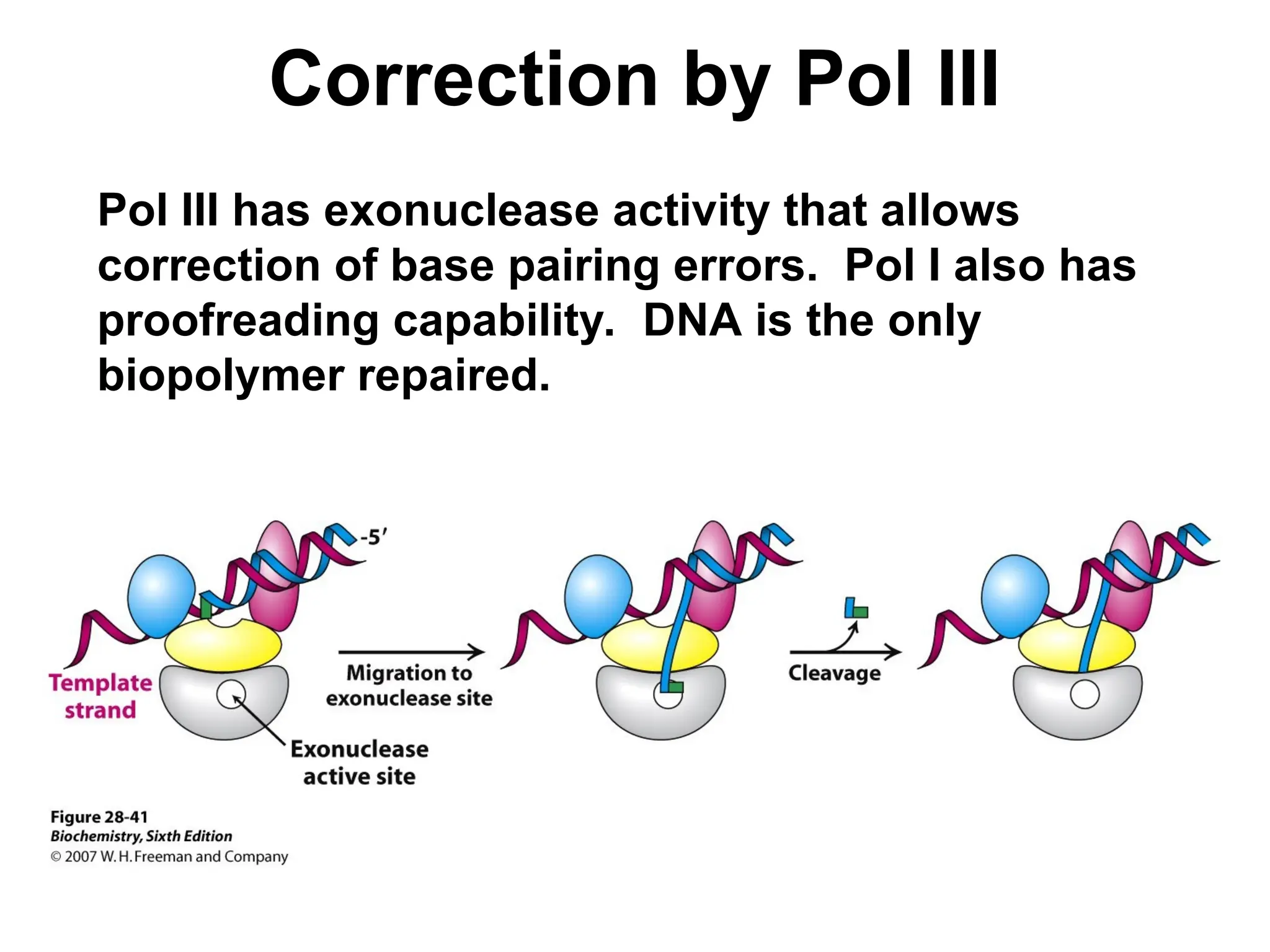
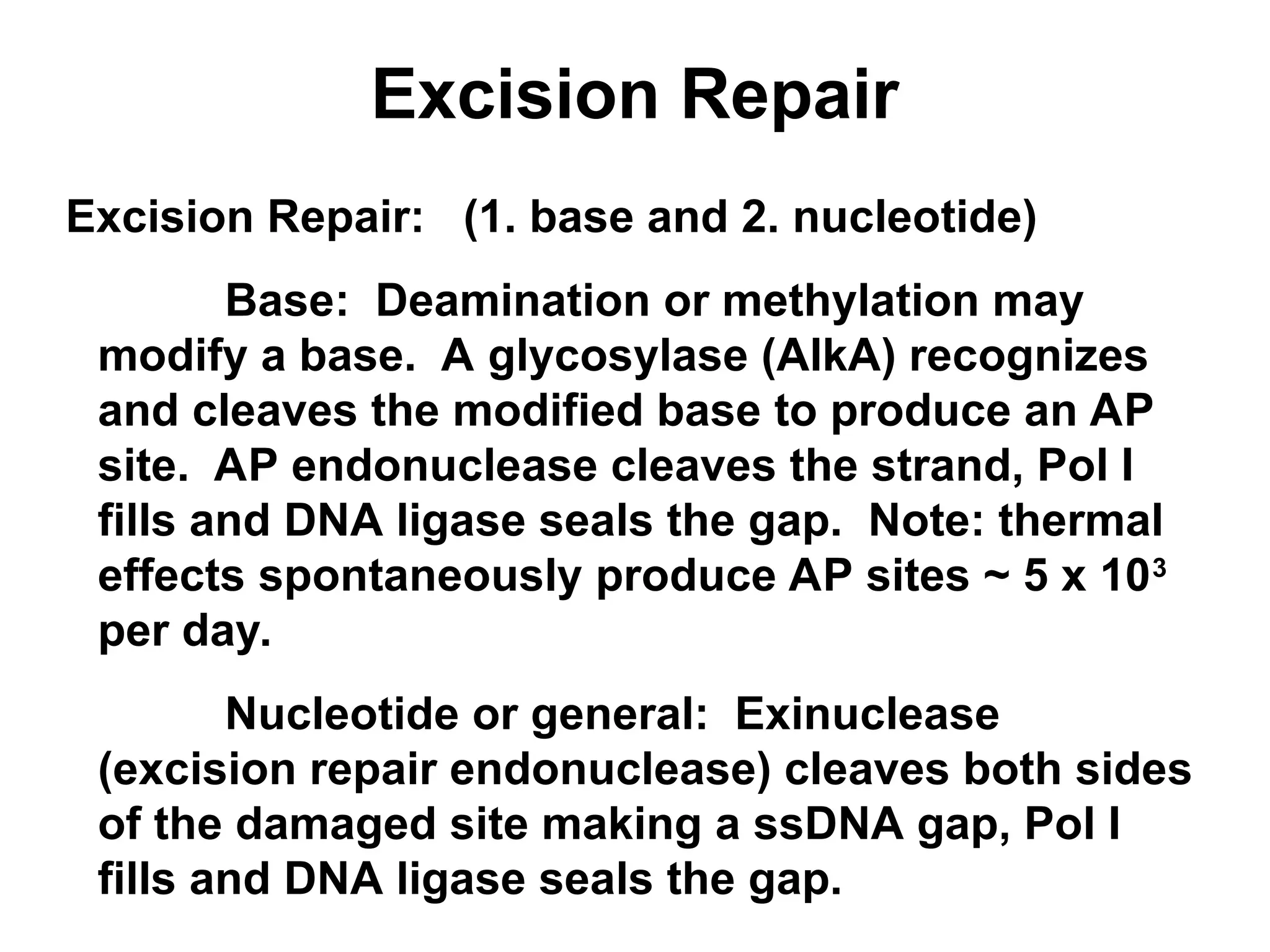
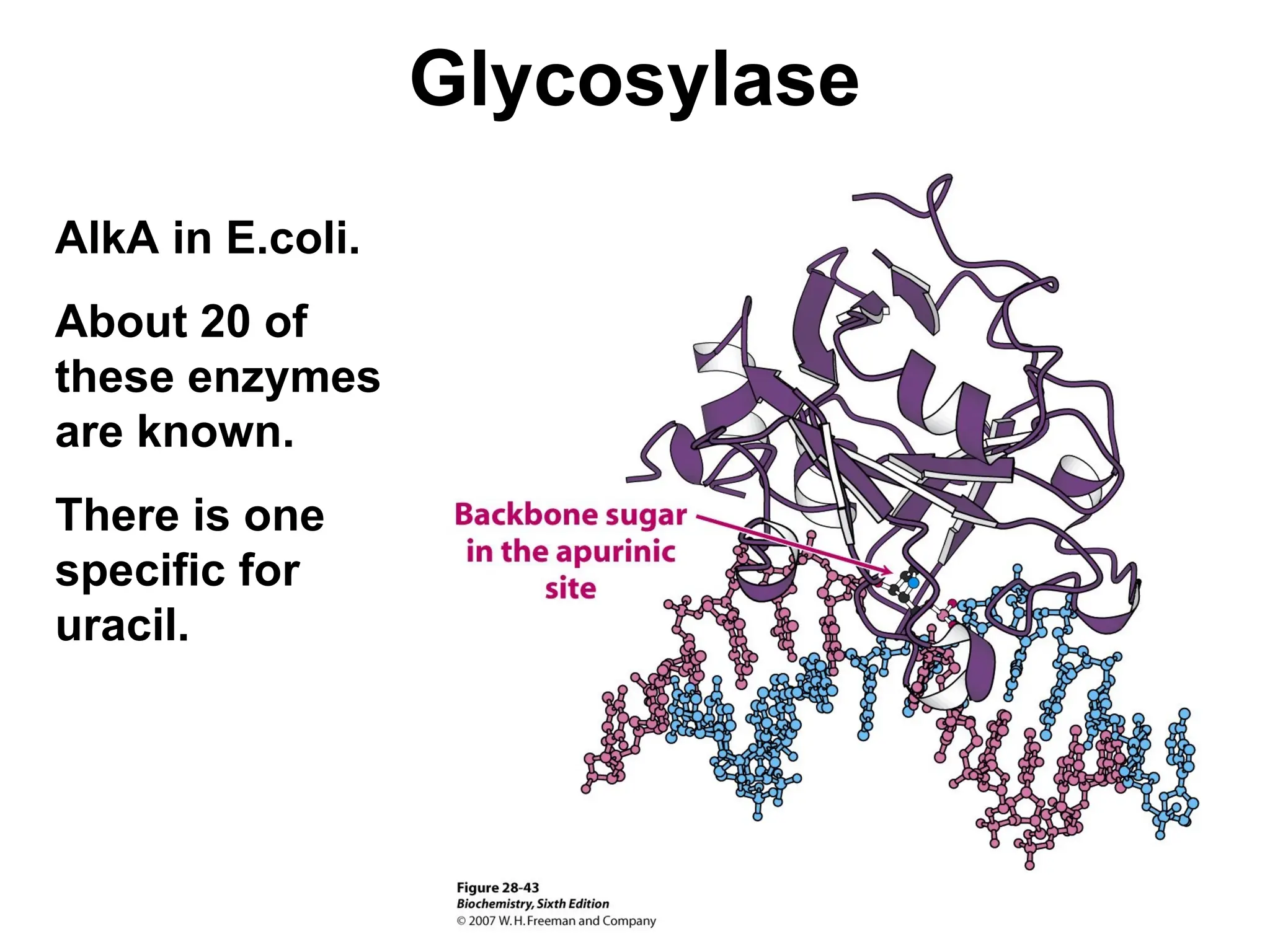
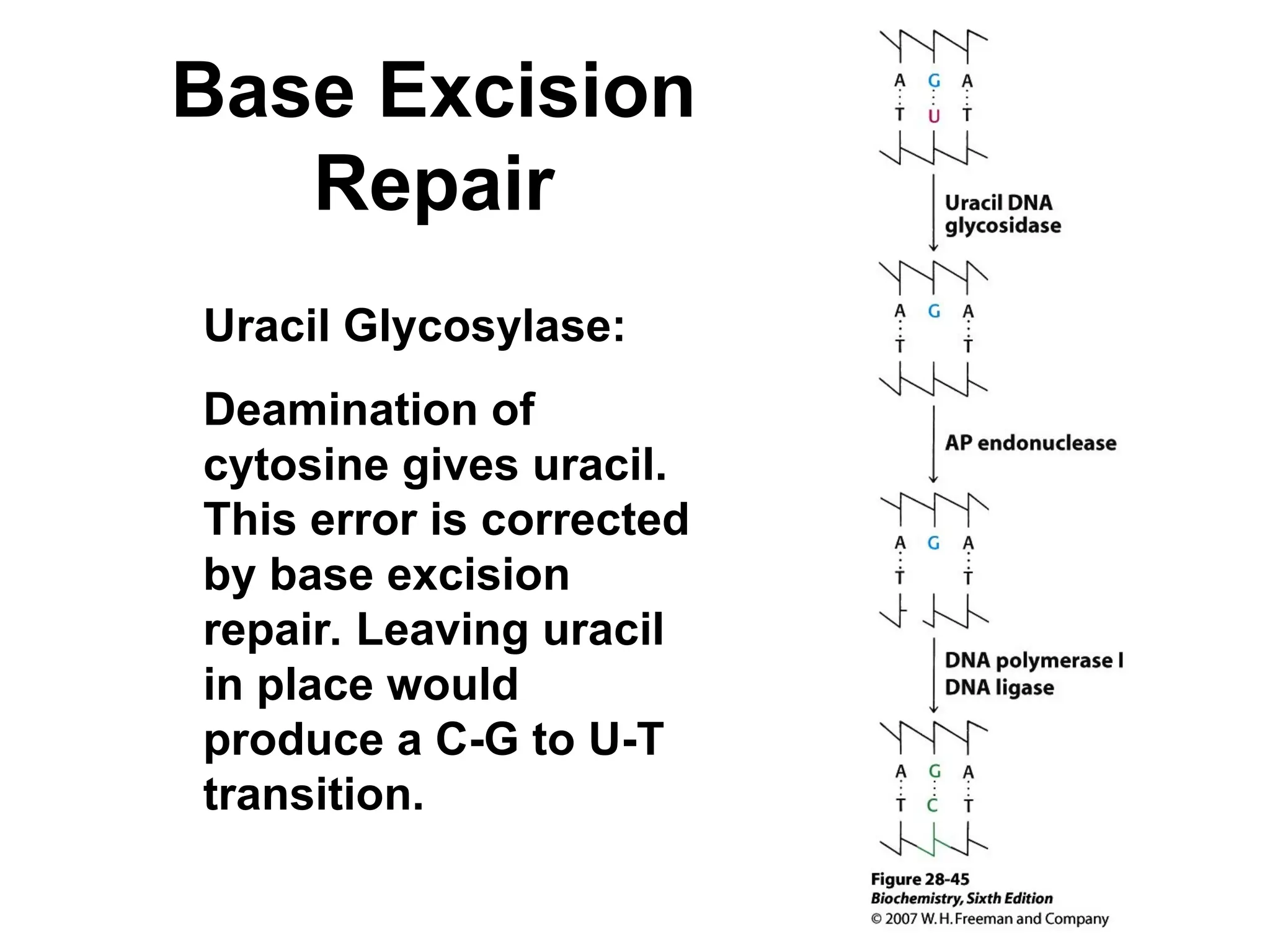
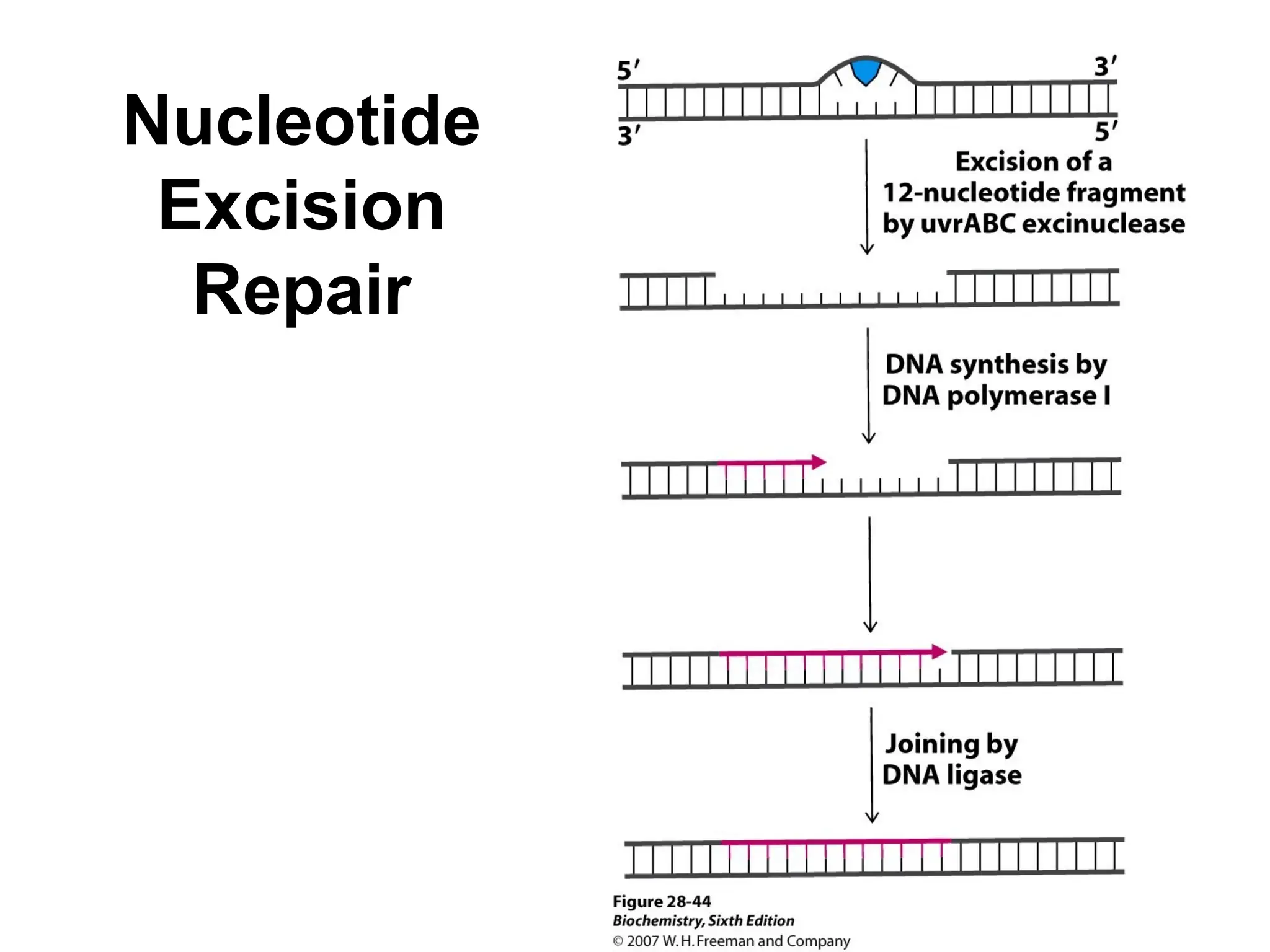
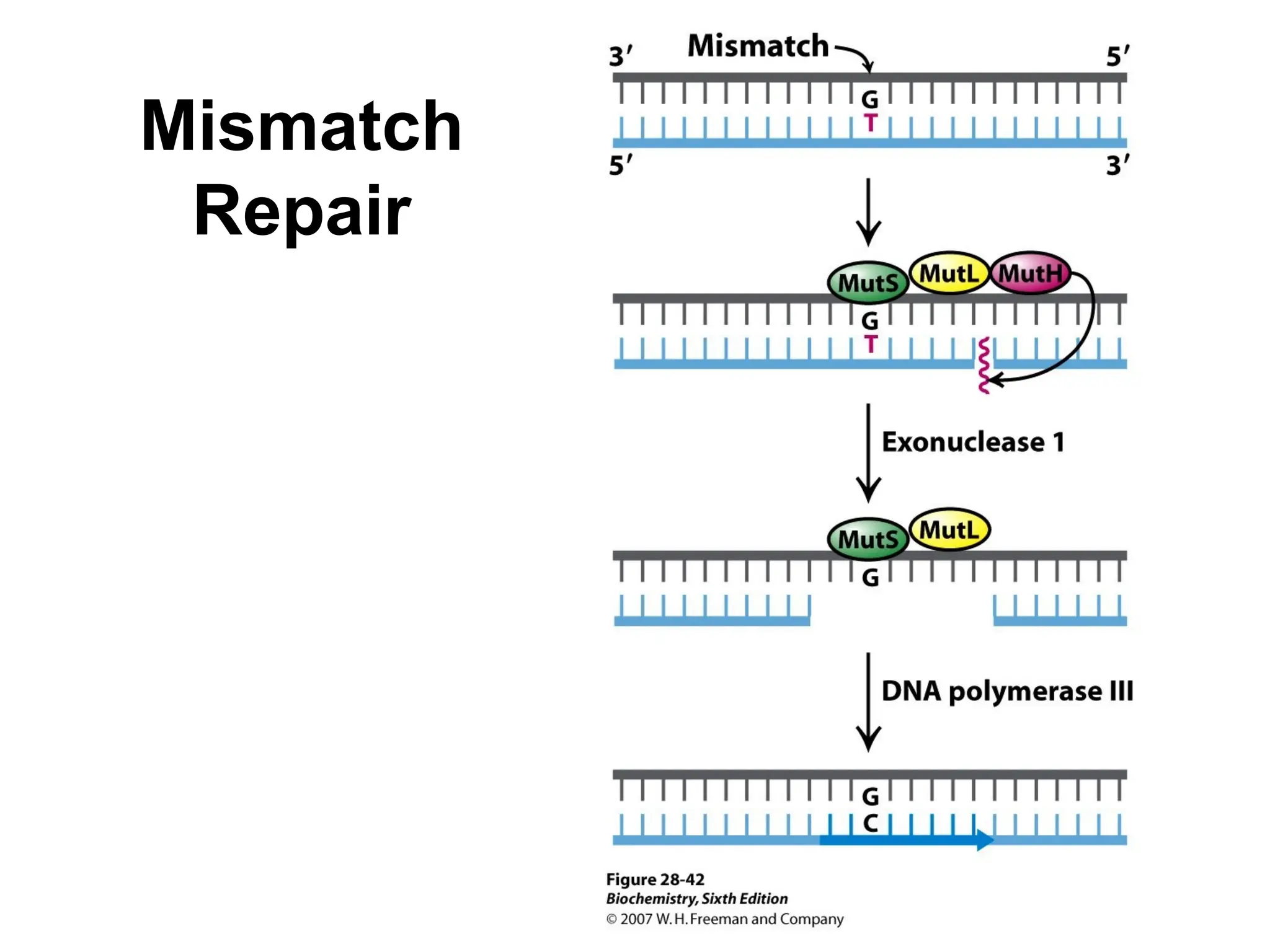
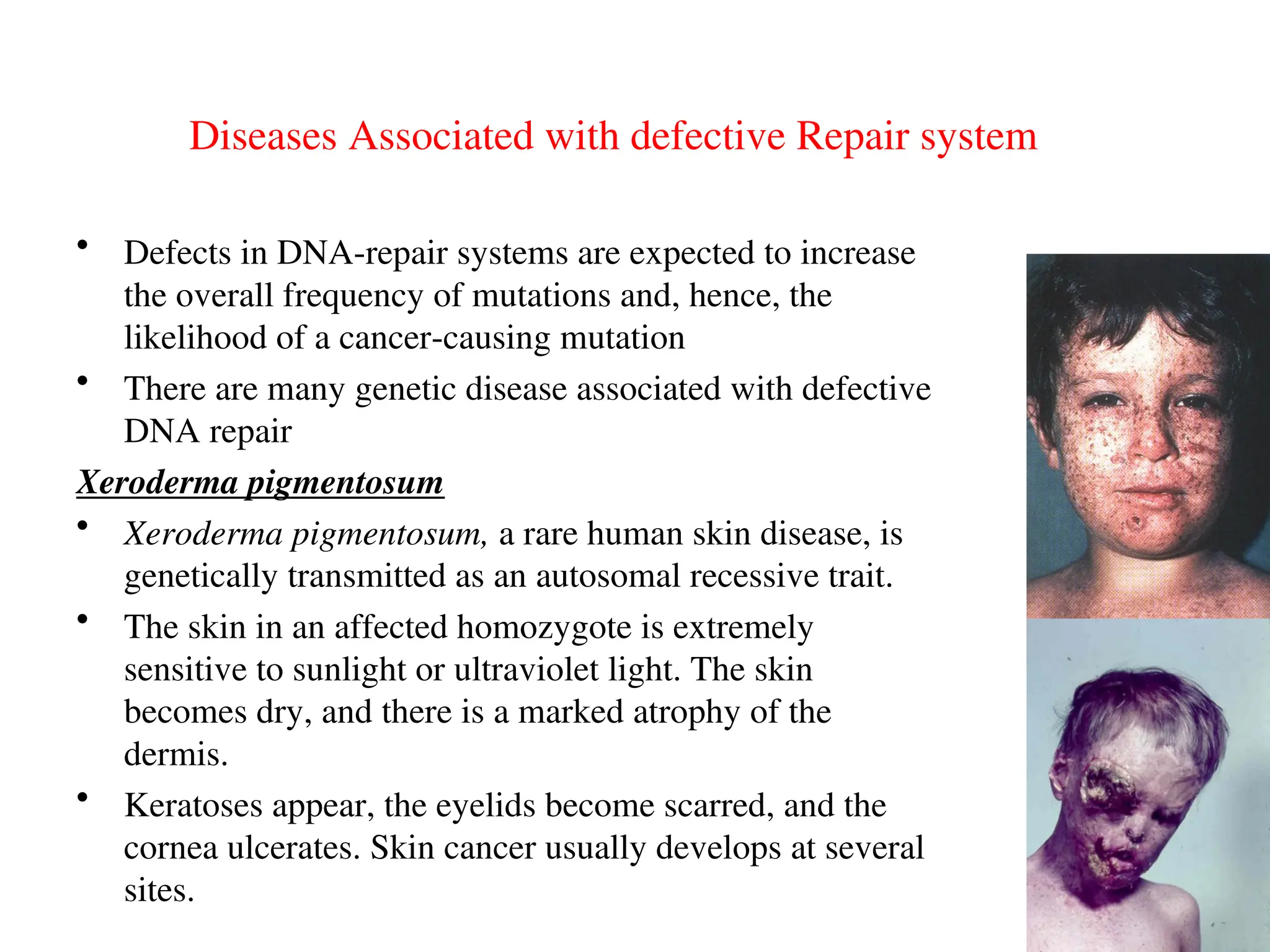
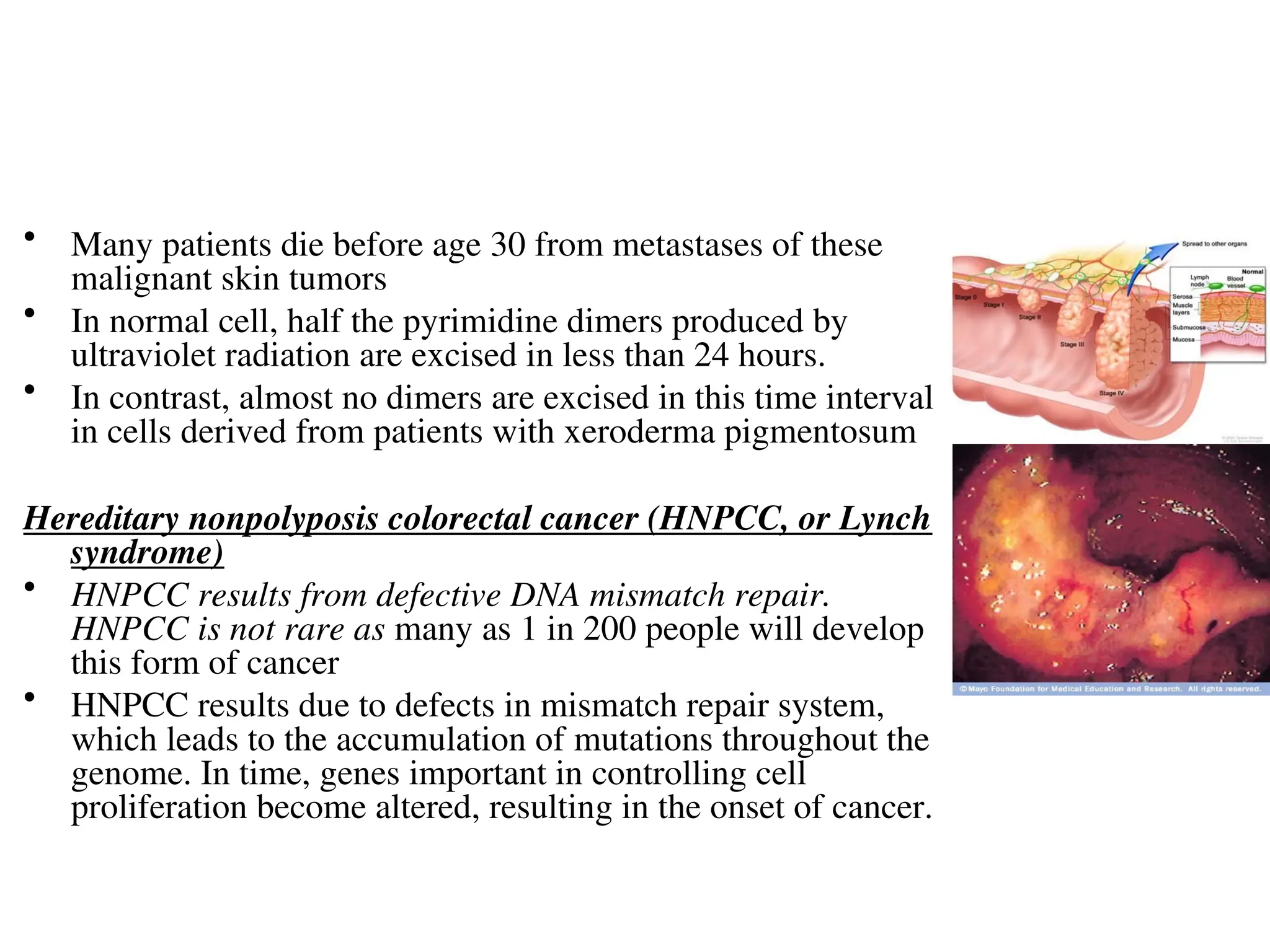
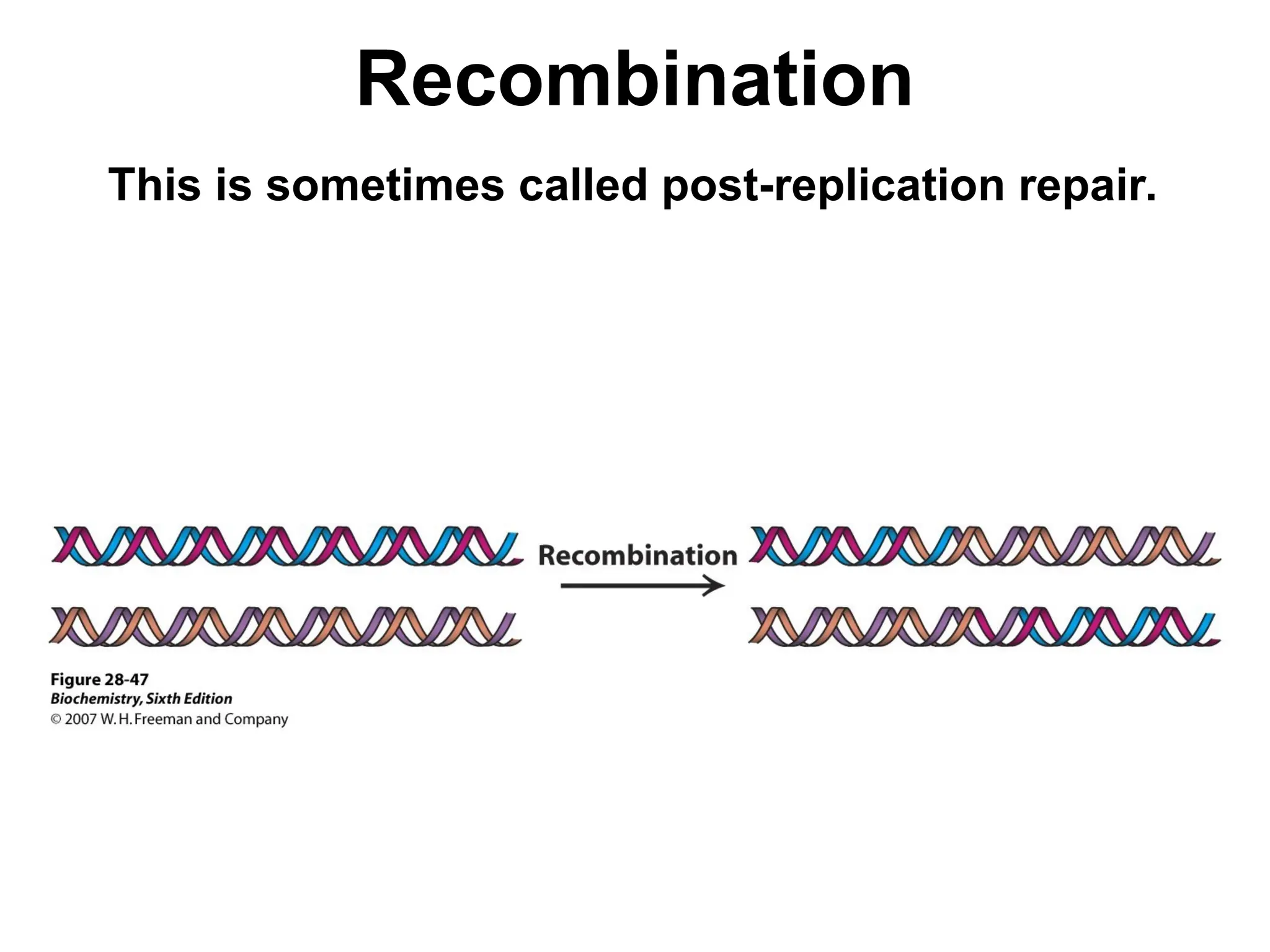
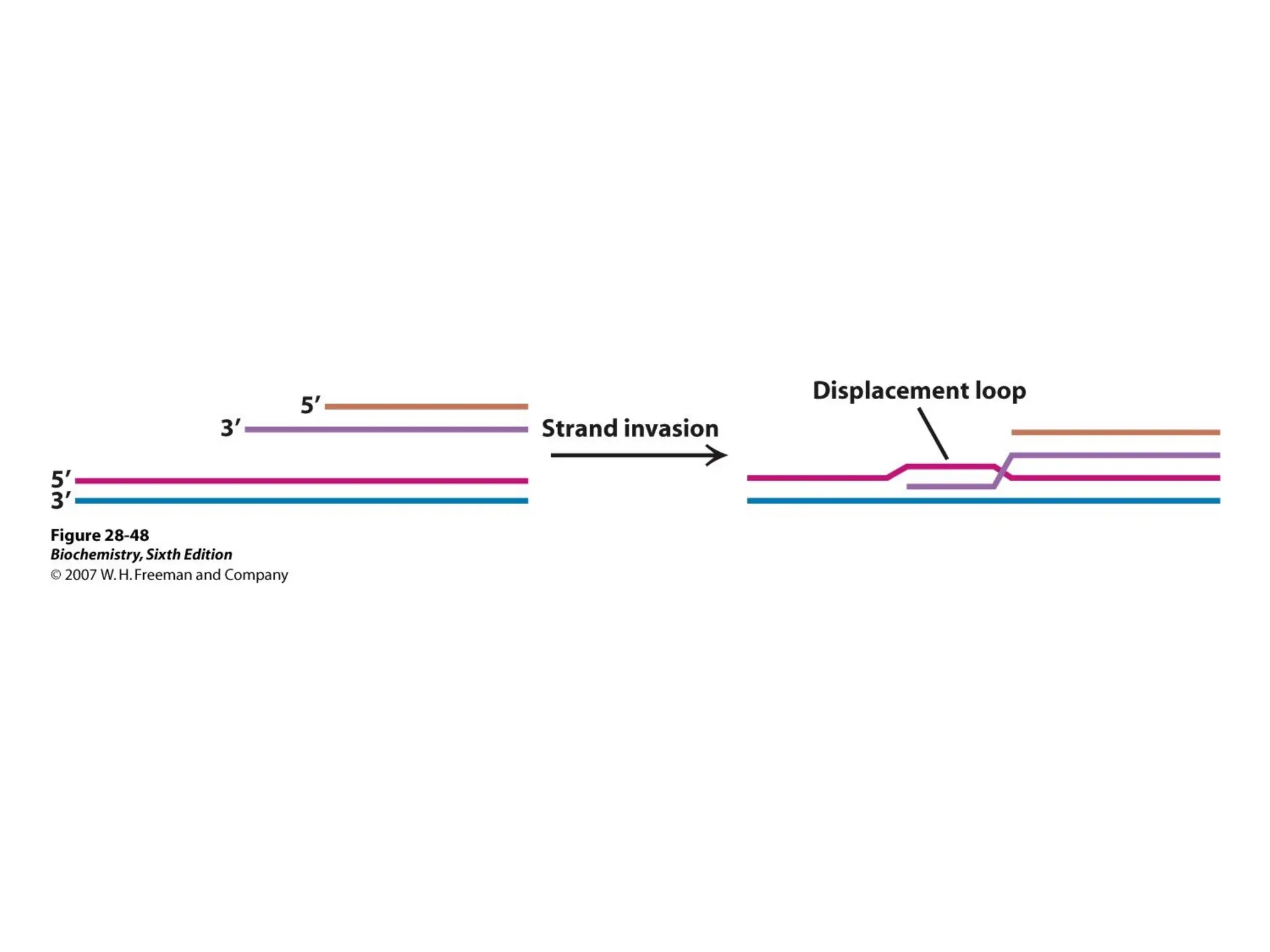
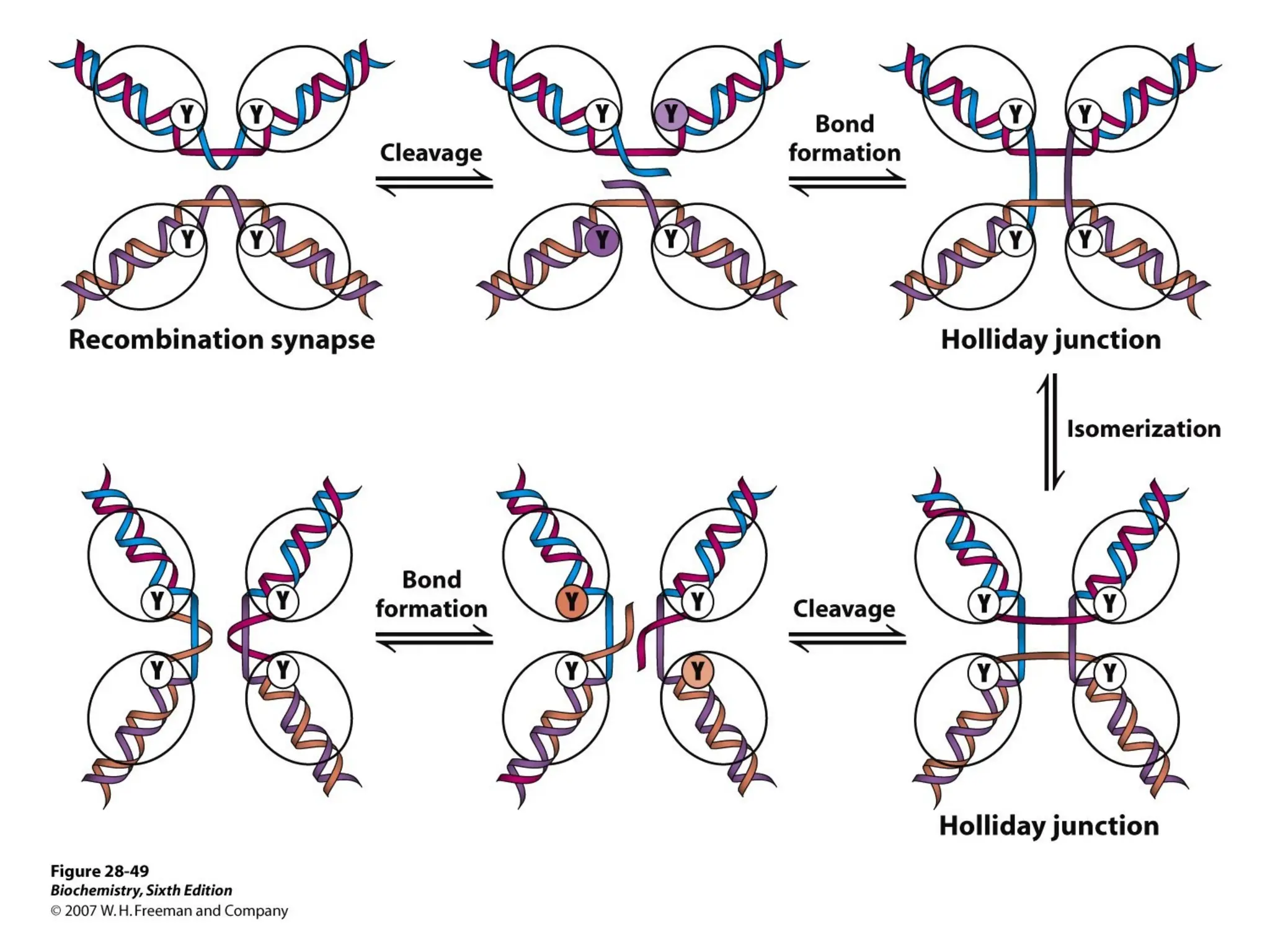
Chapter 28 discusses DNA replication, repair, and recombination, highlighting the accuracy of DNA synthesis along with proofreading mechanisms that reduce error rates. It details the roles of various enzymes, such as DNA polymerases and topoisomerases, in facilitating efficient DNA replication and repairing damages that can lead to mutations or cancer. The chapter also covers the processes involved in priming and completing DNA synthesis along with mechanisms for correcting errors and repairing DNA damage.